Caleb Lareau
@CalebLareau
Followers
4K
Following
12K
Media
196
Statuses
2K
Assistant Professor at Memorial Sloan Kettering. Interested in computational and translational immunology. crazy cat guy. Same handle at the new place 🦋
New York, New York
Joined October 2015
Some big news: I’m absolutely delighted to announce that I will be starting my group this fall in the Computational and Systems Biology Program at @MSKCancerCenter in New York City! 1/.
92
76
739
Encephalitis caused by human herpesvirus 6 has been repeatedly observed in CAR-T cell case studies and clinical trials. In a new work with @satpathology, we make the surprising observation that the cell therapy itself may be the source of lytic virus. 1/n.
13
166
616
On bioRxiv today, we raise serious concerns over a recent @nature paper describing ReDeeM, a new single-cell mitochondrial DNA (mtDNA) sequencing / analysis method. Here are three things that you need to know: 1/n.
7
122
521
Out now in @NatureBiotech, ASAP-seq and DOGMA-seq!! Together with @emimitou, @psmibert, Kelvin Chen, Shimon Sakaguchi, @LeifLudwig, @bloodgenes, Aviv Regev, and many others, we are thrilled to share out the latest in single-cell multi-omics!.
13
140
499
What a PERFFect day to share our new preprint!. With @tsion_abay #BobStickels @MerilTakizawa @ChaligneRonan and @Satpathology, we introduce PERFF-seq, a new experimental approach to studying rare cells with scRNA-seq via transcript-specific enrichment. 1/n.
22
155
466
Out today in @Nature, we describe HHV-6 reactivation in T cells, including therapeutic CAR T cells. I’m so grateful to have worked with an all-star team during my time in the @Satpathology lab that led to these findings. 1/n.
21
121
459
Out today in @NatureGenet, our work utilizing single-cell multi-omics to define specific immune cell subsets that experience purifying selection against pathogenic mitochondrial DNA. Check out a thread about this work below: 1/22.
13
78
312
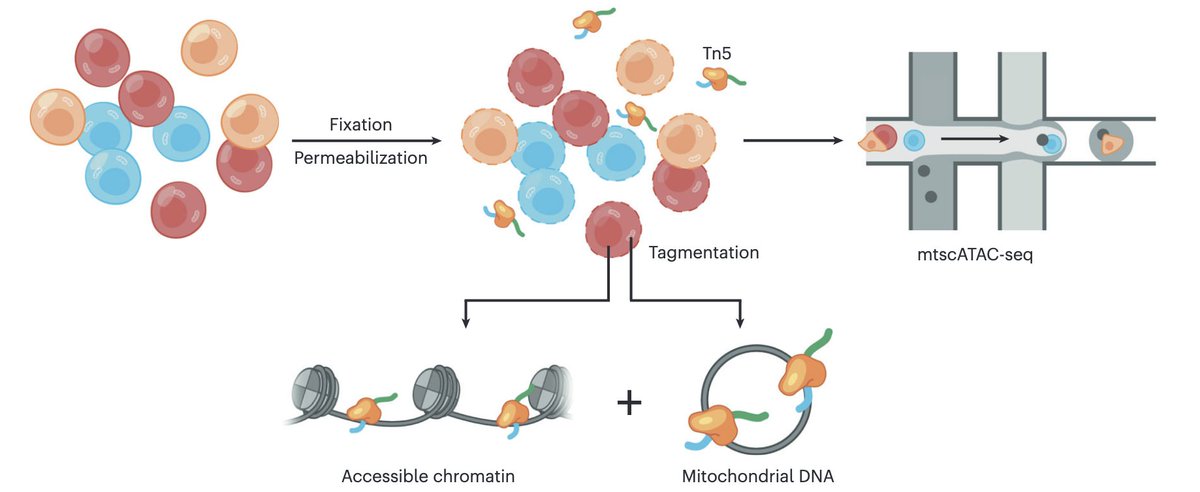
Out today in @NatureProtocols, our detailed writeup on the mitochondrial scATAC-seq assay, spanning everything from the molecular biology to computational analyses. Check out the paper here: and some highlights in this thread: (1/11)
5
53
278
It's #mito mania today in @NatureBiotech @NEJM. The fruits of great collaborative work lead by @LeifLudwig @MWalkerMDPhD @bloodgenes @VamsiMootha and Aviv Regev 1/n.
9
86
236
A common assumption in #singlecell data is that 1 cell = 1 barcode. You've heard of cell doublets, but what about barcode doublets? In work with the great team of Sai Ma, @fabianamduarte, and @JD_Buenrostro, we quantify this effect: Thread: 1/n.
1
97
211
PERFF-seq is out today in @NatureGenet! Our recent thread summarizes why we are excited about the prospects of programmable nucleic acid cytometry for studying rare cell states.
What a PERFFect day to share our new preprint!. With @tsion_abay #BobStickels @MerilTakizawa @ChaligneRonan and @Satpathology, we introduce PERFF-seq, a new experimental approach to studying rare cells with scRNA-seq via transcript-specific enrichment. 1/n.
10
47
202
Out on @biorxivpreprint, I'm excited to share our latest deep dive into the world of mitochondrial genetics using single-cell genomics. Grateful to have worked with @LeifLudwig, @Satpathology, and many others. Thread here: 1/n.
7
44
185
Wow! I am honored to be in the company of amazing #ForbesUnder30 honorees. I could not have made it this far without the support of awesome mentors, peers, and friends. Check out our awesome cohort here:
33
13
167
A one-tweet summary of our new work out now in @nature led by @JulesGrunewald @RonghaoZhou @sarapintogarcia @becauseBiology w/ M. Aryee & JK Joung. Figure 1: CRISPR is broken!!!.Figure 2: CRISPR is saved!!!.Figure 3: CRISPR is broken!!!. Read more here:
3
58
156
Some suggested weekend reading: a new primer out in @TrendsinBiotech from our first postdoc @arthurwchow on key concepts and new developments in single-cell multi-omics! Check it out here!
1
35
148
Come be my colleague! . We are hiring a faculty member in the Computational and Systems Biology Program @MSKCancerCenter. If you are on the market and want to learn more about MSK or the csBio program, I’m happy to share my wonderful experiences here! .
2
64
133
Update on the clonality analysis: now with TCRs! Thanks to @BenceDaniel2, @InflammatoryDC, and @Satpathology for the push. I'm seeing a massive clonal bottle beck in the data. At around immunization 9, we start to see a single TCR-B take over at ~70% of the population.
Very inspired by this paper-- I don't think I've ever seen anything like this. I've never been able to lineage trace (with mtDNA) mutations any system over this time span, so I took a peak at the GEO data and found something potentially interesting? 1/n.
9
22
127
Thrilled to have worked with Kevin Parker and @Satpathology on our new commentary out in @Cancer_Cell where we discuss the state and the future of targeted therapies in fighting cancer-- A short thread on our ideas:.
3
41
125
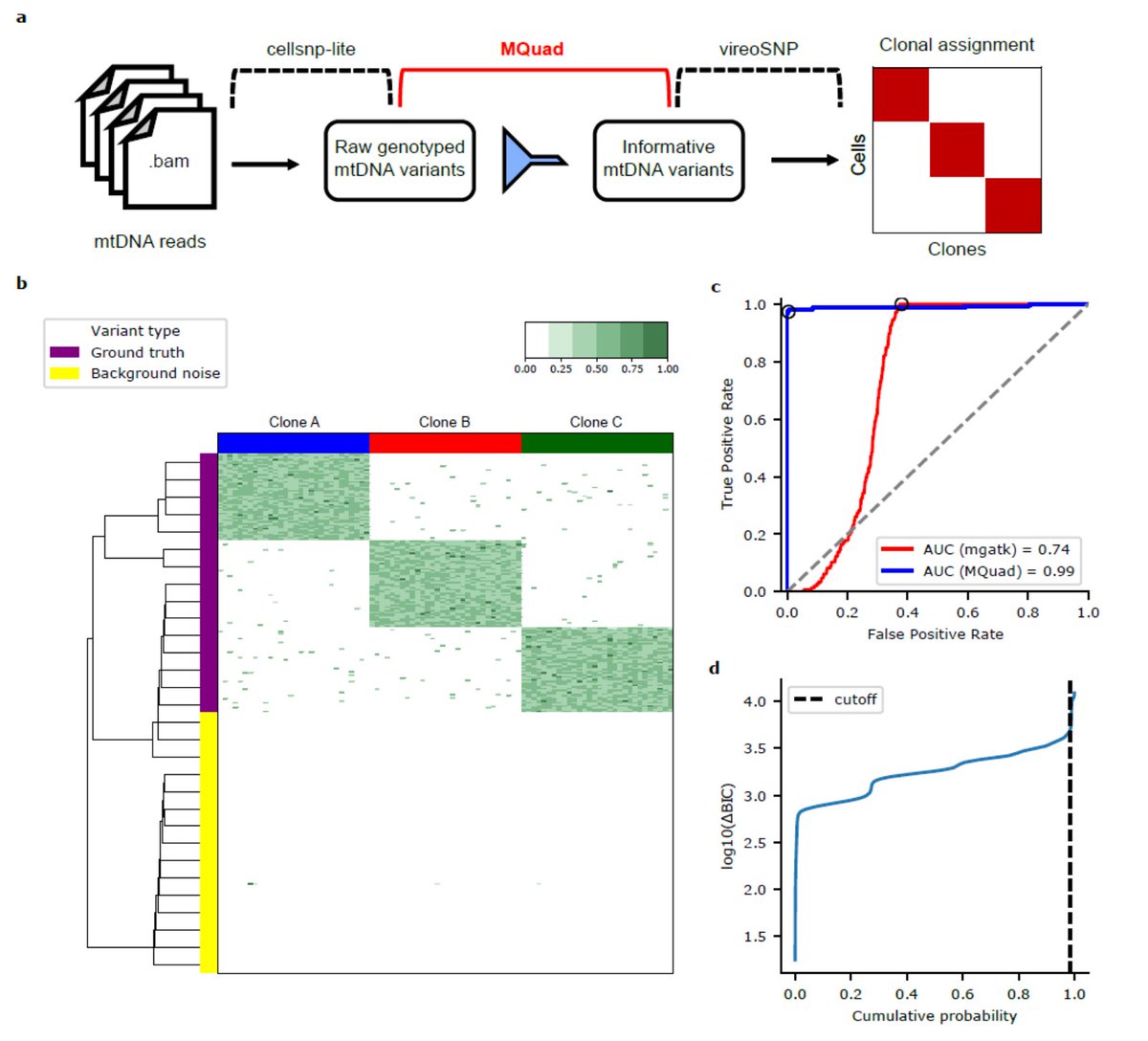
I'm genuinely excited to see the extension of methods to discern clonal structure from mtDNA variants-- something that I've thought about over the years. However, I'm going to comment on a couple of points in this pre-print to hopefully show why mtDNA tracing isn't easy 1/n.
MQuad preprint is up. It effectively detects mitochondrial variations for clonality analysis. We saw mtDNA variants are often complementary to nuclear SNVs or CNVs. Check it out with your scRNA, scDNA or scATAC data.
3
23
118
Our work characterizing barcode multiplets is now out in @NatureComms. Fantastic work with Sai Ma, @fabianamduarte, and @JD_Buenrostro that really benefitted from some excellent, careful peer-review. 1/8.
6
42
103
So pumped to be out of stealth 🥷 Cartography has an exceptional vision for the future of immunotherapies and an all world team to execute! Checkout our launch here! @satpathology @krparker @HowardYChang @MaxMumbach @JeffreyVerboon.
We’re excited to officially launch Cartography Biosciences today with $57M in initial financing. With this support, we will create a smarter roadmap for #cancer immunotherapies with our precision antigen expression atlas. 🧵. #Oncology #MedTwitter.
1
10
102
A massive thank you to everyone for their feedback on the commentary, particularly those who have followed up about their experiences with their own ReDeeM data. I want to add a couple of forward-looking points: 1/n.
On bioRxiv today, we raise serious concerns over a recent @nature paper describing ReDeeM, a new single-cell mitochondrial DNA (mtDNA) sequencing / analysis method. Here are three things that you need to know: 1/n.
1
10
99
Impressive work from the @satijalab introducing Seurat v4! That said, I'm even more excited for Seurat v14, where an entire human cell atlas will almost certainly (r^2 = 0.96) be introduced with the publication. I have no doubt that @NYGCtech is up to the task!
We are excited to release:.* Our new preprint on weighted nearest neighbor analysis to define cell states based on multiple modalities in Seurat v4.* A multimodal CITE-seq dataset of human PBMC w/200k cells and 228 antibodies .Data/code are available now!.
2
7
89
Delighted to highlight SEACells-- a fantastic new method from the labs of @SettyM and @dana_peer-- in @NatureBiotech today. Work smarter, not harder, with your #scATACseq and #scRNAseq data.
@MSKCancerCenter @ColumbiaCompSci @CUSEAS @ColumbiaScience @fredhutch @HHMINEWS @SettyM @dana_peer Subtle cell states resolved in single-cell data - @CalebLareau #NBTNV
2
18
86
Are you interested in completing your postdoctoral work at @Stanford? We are hosting a Stanford Science Fellows webinar from noon-1pm on September 9th-- come hear about the program and get your burning questions answered!
2
44
83
Big innovations in scATAC-seq out in pre-print today. First, our paper with @fabianamduarte, Jen Chew, Vinay Kartha, and others at @BioRad and advised by @JD_Buenrostro describes significant technical innovations in data quality, cell capture, read abundance, and informatics. 1/4.
2
33
83
Thank you @chenweng1991, @jswlab, @bloodgenes for the feedback on our work. @LeifLudwig and I agree that the “-2” filtering method that mitigates edge bias is a critical development for ReDeeM. Nevertheless, we stand by our original preprint, noting the following: 1/n.
1
14
73
Thrilled to see this collaborative work out in @SciImmunology led by @JessicaWang927 and @gardnerlabUCSF at @UCSFDC and Stanford in the @Satpathology lab.
4
17
67
We're hiring at @StanfordPath! With research support from @parkerici, @genome_gov, and @Stanford Science Fellows, we are looking for a junior computational biologist to learn single-cell multi-omic analysis directly from me in the @Satpathology lab
1
35
68
Thank you @statnews for the incredible honor and for the opportunity to attend #STATSummit. I'm so excited to meet the other #STATWunderkind honorees this week in Boston!.
It's here! Our sixth class of @statnews Wunderkinds, those early-career researchers and doctors already making waves and headed for even bigger things.
9
4
55
Our lab will be hiring at all levels for both experimental and computational positions! Please reach out or find me at #HCA2023GM if you have an interest in joining, collaborating, or have any advice for a new PI! 5/fin.
7
19
52
The team of @emimitou and @psmibert make it look easy. This assay went from idea to proof of principle to biological insights in a matter of weeks. unreal efficiency and innovation from @NYGCtech 1/6.
Excited to share our latest single cell multimodal assay. ASAP-seq pairs surface and intracellular protein detection with high quality chromatin accessibility on the @10xGenomics #scATACseq platform.
1
6
39
Thank you to the exceptional colleagues that shaped my scientific journey, esp. @satpathology and @anshulkundaje for opening their labs to me at Stanford; @bloodgenes, @jd_buenrostro, and @LeifLudwig for training me in my PhD. It’s exhilarating to join their ranks as a PI. 4/
1
1
34
We are hiring a Life Sciences Research Professional here at @StanfordMed! Come work with me and @Satpathology to apply cutting edge single-cell genomics assays to understand fundamental properties of disease!
0
17
32
A tour-de-force effort from (Dr.!) @TimoRckert and @RomagnaniLab characterizing (clonal) adaptive immunity of NK cells IN HUMANS! So happy to have worked with this team on this fascinating problem. Check out this thread to see how single-cell multi-omics enabled these🔥insights.
We are excited to share our recent paper with the title “Clonal expansion and epigenetic inheritance shape long-lasting NK cell memory”, led by @TimoRckert, in which we study the origin and maintenance of human adaptive NK cells!
0
4
30
First there was spatial without imaging… Now there is single cell without sequencing…
STAMP is OUT!. Sequencing-free #singlecell RNA and Protein profiling. Game-changing approach for scalable and cost-efficient analysis of single cells. The future starts today!. Amazing @LGMartelotto @DrJasPlummer.
0
6
29
I’m incredibly thankful for the friends who shaped these analyses and interpretations: @doctor_msc, @livius_tacitus, @nawy_t, @dana_peer, and @LeifLudwig. Feedback is very much welcomed! 19/n.
0
0
32
Check out this awesome digest on PERFF-seq from @tsion_abay about our time developing it in the @Satpathology lab!.
Work with rare or difficult to sort cells? Don’t miss this blog post! Learn about PERFF-seq, a new method for RNA-based enrichment of cells prior to scRNA-seq with Chromium Flex, and read valuable insights from method co-developer @tsion_abay.
1
3
27
@10xGenomics Thanks for the highlight! mtscATAC-seq was co-lead by @LeifLudwig while we were at @broadinstitute @BostonChildrens @harvardmed working with Aviv Regev and @bloodgenes. It takes a whole team to hack a problem like this!
2
3
27
@anshulkundaje @vallens @RongxinFang Currently void of content, but now in progress. Database (comment only; please DM if anyone wants to help curate): Submission form (public):
3
12
25
Awesome to see our work on mtDNA lineage tracing highlighted at @satijalab’s #singlecellgenomicsday. Tune in to the live stream here:
1
0
25
If this paradigm appeals to you-- massive-scale data analyses to motivate experiments for new discoveries in cancer immunotherapy, my brand-new group at @MSKCancerCenter is hiring to pursue other questions in this framework! Please get in touch! 11/n.
0
4
22
I use this workflow all the time-- for everything from gRNAs to antibody barcodes. It was instrumental in getting our ASAP/DOGMA-seq preprocessing workflows up and running quickly. Glad to see the preprint out!.
In a new preprint, @lioscro, @JaseGehring, @lpachter and I describe a method and software (kITE) for quantifying orthogonal barcodes from assays such as Perturb-seq, Clicktagging, TAPSeq, CiteSeq, Multiseq, and 10xFeature Barcoding: 1/
0
6
19
Curious about PERFF-seq and how it can be helpful for your research? Come check out @tsion_abay's webinar tomorrow (6/18) at 9am PT / noon ET! 🐟🐟🐟🐟🐟🐟🐟.
What a PERFFect day to share our new preprint!. With @tsion_abay #BobStickels @MerilTakizawa @ChaligneRonan and @Satpathology, we introduce PERFF-seq, a new experimental approach to studying rare cells with scRNA-seq via transcript-specific enrichment. 1/n.
0
7
23
In any case-- very cool data @Masopust_Vezys -- thanks for uploading your sequencing to GEO to enable niche analyses like these!.
0
0
22
@JeanMolinier2 @MicrobiomDigest @KamounLab @PubPeer It appears that the editor in chief of the journal is a co-author on the study fwiw.
1
1
19
What’s even better than a tweet thread? A YouTube live stream! Come learn about PERFF-seq, including assay development and performance, TOMORROW (3/29) at #singlecellgenomicsday hosted by.@satijalab.(my talk is at noon ET): 19/n.
1
5
19
I moved across the country during a pandemic to start my fellowship at @Stanford. It's extremely disheartening to see faculty like @MLevitt_NP2013 and affiliates like @SWAtlasHoover use their platform at this university to push dangerous, un-scientific nonsense. What can be done?.
How did emails sent to @ISCB_info 14 Mar. launch Lior Pachter @lpachter on his campaign of anti-science hate? . Must be his fear! He needs to stop being so scared. It is not good for him or those who believe a word he says. DO THINK as if your life depends on it. IT DOES!
0
0
17
I want to give a special shout out to my wonderful colleagues to made this work happen, including @majzner_lab, @satpathology, and others not on Twitter. Their boundless energy and creativity made assembling this story a very fulfilling experience. 18/n.
1
0
19
🔥🔥🔥🔥🔥 from @maple1989. Amazing, tireless execution of a plan and a vision that's been years in the making. Awesome work!!!!
Our SHARE-seq paper for single-cell ATAC+RNA co-profiling is now in Cell! .@CellCellPress .
3
1
19
@anshulkundaje @NatureBiotech @emimitou @psmibert @LeifLudwig @bloodgenes "uh oh, back to the lab again. "
0
3
16
The most critical data came in collaboration with an awesome teamwork with friends at @DanaFarber and @StJude. Here, we show that we can identify HHV-6B+, CAR+ T cells in vivo from patients receiving clinical and FDA-approved CD19 CAR-T cells. 12/n
1
0
16
Congrats to Dr. @isabelfulcher for successfully defending her dissertation today. Thanks for setting the pace in the relationship #PhD #stayinschool #harvardbutchill
2
2
15
Congrats @EMC22381830 👏 !!!! . And thanks to @MSKEducation for sponsoring such a great week for our post docs!.
🥇Daniella Audi Blotta, MD (@DABlotta) from the @MarkRobsonMD lab.🥈@VittoriaBocchi, PhD from the @studerl lab.🥉@CarlinLiao, PhD from the @nadeem_lab . 🏆people's choice goes to Erin Cumming, PhD (@EMC22381830) from the @CalebLareau lab.
0
1
18
Given the striking reactivation of HHV-6 in standard T cell cultures, we wondered whether HHV-6 reactivation was occurring during CAR-T culture. With terrific support from friends at @AllogeneTx , we studied the dynamics of HHV-6 reactivation in research CAR T cells. 7/n.
1
0
16
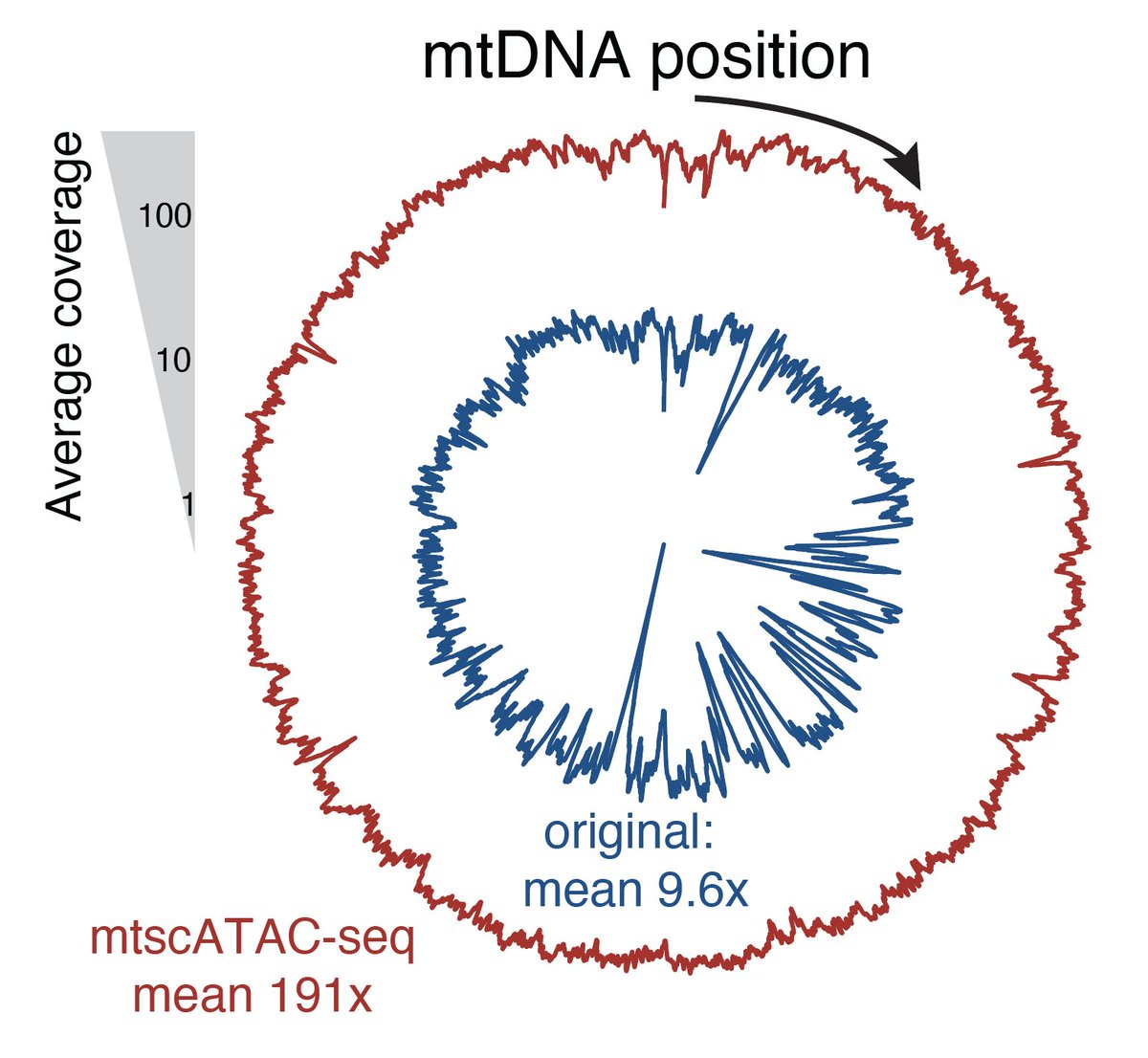
First, @LeifLudwig and Christoph Muus figured out how to retain mtDNA in droplet-based scATAC, which we term mtscATAC-seq. This enabled scalable solutions for performing human lineage tracing using somatic mtDNA variation 2/n
2
2
15
The opportunity to join the world-class science and phenomenal collaborators at @MSKCancerCenter is incredibly special. I am so grateful to find my academic home in csBio, a community of many of the individuals in science that I admire and respect most, led by @dana_peer. 3/.
1
0
16