Rahul Satija
@satijalab
Followers
28,409
Following
383
Media
168
Statuses
900
Core Member, New York Genome Center; Associate Professor, Biology, NYU
Joined September 2009
Don't wanna be here?
Send us removal request.
Explore trending content on Musk Viewer
Vance
• 1094644 Tweets
#เทควันโด
• 527418 Tweets
Iraq
• 436103 Tweets
Stolen Valor
• 274576 Tweets
ETERNITY WITH BLACKPINK
• 241461 Tweets
Wisconsin
• 222495 Tweets
Kursk
• 217104 Tweets
BEST WISHES FOR SAROCHA
• 209892 Tweets
Trump 2024
• 158503 Tweets
#erişimengelinikaldır
• 131119 Tweets
ISIS
• 126047 Tweets
Tampon Tim
• 124170 Tweets
Eau Claire
• 91023 Tweets
Walthamstow
• 66826 Tweets
Zubimendi
• 57205 Tweets
Vienna
• 56844 Tweets
Brighton
• 50818 Tweets
Quincy Hall
• 40104 Tweets
Chidimma
• 36121 Tweets
Home Affairs
• 35634 Tweets
$XRP
• 33813 Tweets
Nelly
• 32227 Tweets
Bon Iver
• 23563 Tweets
Ferdi
• 23277 Tweets
Stona
• 22833 Tweets
#الاتحاد_انتر_ميلان
• 20274 Tweets
Esra Yıldız Kahraman
• 13604 Tweets
Entourage
• 11975 Tweets
D'Tigress
• 11584 Tweets
FURIALEX EN CARAJO
• 11111 Tweets
Last Seen Profiles
As my lab develops Seurat, I wanted to share some thoughts on the important Seurat and scanpy package selection preprint from
@Josephmrich
/
@lpachter
🧵
11
234
952
We've released a Seurat update with support for imaging-based spatial technologies! Our new vignette shows how to download, analyze, and explore public datasets from
@vizgen_inc
(MERFISH),
@nanostringtech
(SMI), and
@AkoyaBio
(CODEX).
Check it out at:
7
222
839
We've been working on Seurat v4 for almost two years, and are excited to share it with you soon!
In a webinar w/
@10xGenomics
on 10/20, I'll introduce new methods to analyze multimodal single-cell data (i.e. from CITE-seq or the 10x multiome ATAC+RNA kit):
11
262
807
Our paper describing Weighted Nearest Neighbor analysis in Seurat v4 is out in
@CellCellPress
! We introduce a framework to analyze and integrate datasets where multiple data types are simultaneously collected in the same cell:
4
220
773
Have been a bit quiet on twitter as we've been getting to know this little guy - say hello to Avyaan Rao-Satija.
And thanks to my lab (esp
@smith_tracymae
) for the amazing Single Cell Genomics Day quilt!
42
1
716
We’re excited to release a Seurat update with support for Spatial Transcriptomics data! Includes clustering, interactive visualization, and integration with scRNA-seq references. Check out our vignette on
@10xGenomics
Visium data:
5
262
683
We've been working on Seurat v5 for two years and are excited to share it with you soon!
In a webinar w/
@10xGenomics
and
@ScienceMagazine
on 3/29, I'll introduce new methods to analyze spatial/multimodal datasets that scale to tens of millions of cells:
5
167
667
Five years ago we released our first version of Seurat. Since then, its been extraordinary to watch the growth of this field and to be part of an uplifting, open, and collaborative community developing analytical tools for single cell genomics.
Try out 1st software release from my lab at
@nygenome
- Seurat: R toolkit for single cell genomics. Code, tutorials-
http://t.co/7BMGF7wtHn
0
17
26
10
47
640
ISAAC-seq (from
@XiChenUoM
lab) profiles RNA+ATAC from the same cell using the 10x Genomics scATAC-seq kit! Data looks very promising- slightly higher reads/cell compared to 10x multiome (and much cheaper).
3
109
518
We are excited to launch the Center for Integrated Cellular Analysis! As part of the NIH
@genome_gov
CEGS program, we will develop methods to measure and harmonize molecular modalities, spatial context, and lineage history across single cells: (1/3)
15
88
511
Stunning preprint from
@WangXiaoLab
and
@ganoopyliujia
introducing STARmap PLUS for in-situ transcriptome profiling. Used to generate a 3D map of the whole mouse brain and spinal cord (1,000 genes, 1M+ cells)
1
133
496
Two papers in
@CellCellPress
integrating scRNA-seq datasets across modalities and technologies, including CITE-seq, scATAC, methylation, and spatial data!
* LIGER (
@macosko
):
* Our Seurat v3 'anchors' (led by
@timoast
+
@aw_butler
):
4
230
502
Interested in single-cell CRISPR screens for combinatorial perturbations?
Check out CaRPool-seq, where multiple gRNA 'carpool' together on a self-cleavable array.
Now out in
@naturemethods
:
And request our free Starter Kit to try it yourself!
5
89
495
Interested in single cell genomics but need help getting started? Check out the agenda for my lab's free workshop next Friday. Very excited that we will be livestreaming all talks with the support of
@cziscience
! Please RT and spread the word!
7
299
483
Interested in single cell genomics but need help getting started? Check out the agenda for my lab's free workshop next Friday. Very excited that we will be livestreaming all talks with the support of
@cziscience
! Please RT and spread the word!
12
337
477
We are excited to introduce Phospho-seq, our approach for profiling intracellular protein and cell signaling dynamics alongside additional molecular modalities in single cells!
Check out
@JBlairSci
's preprint at
7
121
475
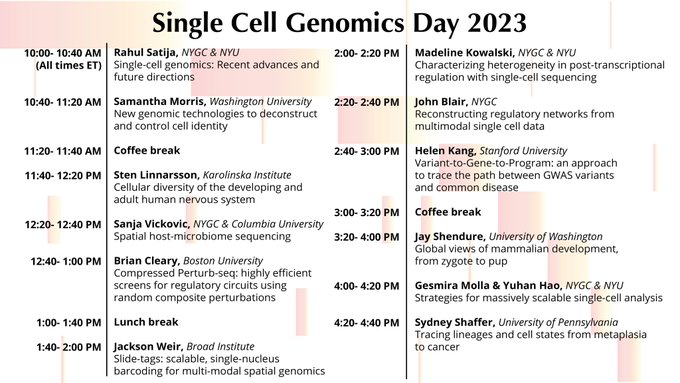
Want to hear about the latest updates/new techniques in single cell and spatial genomics? Check out my lab's eighth annual Single Cell Genomics Day on March 29. Keynotes from
@IdoAmitLab
@JD_Buenrostro
and Xiaowei Zhuang. Please RT/spread the word!
1
184
463
Our new preprint, led by
@YUHANHAO2
, introduces bridge integration: cross-modality alignment via a multiomic 'bridge' dataset.
We map single-cell ATAC-seq, methylation, CUT&Tag and CyTOF profiles onto scRNA-seq references- scaling to millions of cells.
3
116
466
We've updated our scRNA-seq/scATAC-seq integration vignette to analyze a
@10xGenomics
Multiome dataset (ground truth), and benchmark our ability to perform cross-modality annotation and integration:
1
97
457
Our scCUT&Tag-pro and scChromHMM manuscript is out
@NatureBiotech
! Led by
@Bingjie_Zhang_
and
@k3yavi
, we use surface proteins to integrate data from six histone modifications, RNA and ATAC - and explore chromatin state heterogeneity in single cells:
2
140
437
Interested in single-cell genomics but need help keeping up with a rapidly changing field? Check out our free
#singlecellgenomicsday
workshop, starting in a few minutes! Full agenda and livestream at , or follow along below:
14
134
441
Impressive work from
@hattaca
and Regev lab introducing inCITE-seq: simultaneously measuring intracellular proteins (including TFs!) and gene expression in single cells:
6
130
436
Very exciting work from Bing Ren and
@CEpigenomics
introducing Paired-Tag, for simultaneous transcriptome + CUT&TAG profiling in single cells:
1
132
432
Our review, led by
@timoast
, on Integrative Single Cell Analysis is out
@NatureRevGenet
. We cover:
* Technologies for multimodal profiling
* Analytical methods for data harmonization+joint learning
* Integration of spatial and sequencing datasets
3
214
422
We're excited to release an Azimuth reference of human fetal development. Leverages an incredible sci-RNA-seq3 dataset of 15 organs from
@coletrapnell
@jshendure
and
@junyue_cao
.
You can map and annotate your own data onto this reference at
3
99
422
We compared six commercial + academic multiplexed in-situ gene expression profiling technologies. Check out our preprint, led by
@AustinMHartman
.
Takeaway when evaluating spatial tech: consider both sensitivity and specificity!
4
125
411
Two papers using multimodal single-cell CRISPR screens to study immune checkpoints out in
@NatureGenet
!
* Perturb-CITE-seq from Aviv Regev and
@BizarMd
:
* Our ECCITE-seq study identifying PD-L1 regulators:
5
128
416
Excited to share our Seurat update supporting VisiumHD data from
@10xGenomics
. We explore and validate the ability of HD to map the spatial localization of individual cell types:
3
95
406
Three recent manuscripts reporting multimodal (RNA+protein) extensions of spatial transcriptomics!
1. SM-omics (
@mssanjavickovic
) :
2. SPOTS (
@landau_lab
):
3. Spatial-CITE-seq (
@RongFan8
):
0
123
400
Ultra-high throughput scRNA-seq from Christoph Bock's lab! Combines combinatorial barcoding with droplet microfluidics to sequence >150,000 cells on a single
@10xGenomics
lane:
0
138
396
Our free Single Cell Genomics Day livestream starts tomorrow at 10AM EST! You can ask questions during talks via twitter (include
#singlecellgenomicsday
) and we'll relay them to speakers, see you soon!
8
124
388
That's a wrap for
#singlecellgenomicsday
2023! Thanks to our amazing speakers, and to all of you for tuning in!
Its a privilege to celebrate the state of the field for the 7th year in a row, and we look forward to seeing you next year!
(And thanks
@ATJCagan
for illustrating!)
2
44
384
Frustrated by the sensitivity, scalability, and cost of scRNA-seq? Check out our most recent collaboration w/
@NYGCtech
: Robust, doublet-free, and low-cost molecular profiling of biological systems:
13
87
371
Takeaways from amazing
#KSsinglecell
:
1. Spatial technologies are mature, and are tremendously exciting
2. Integrating scRNA-seq data with cell morphology, location, and functional readouts is key for interpretation
3. Single cell genomics is transforming developmental biology
4
121
364
Want to infer causal regulatory changes from scRNA-seq/spatial data? We combined Perturb-seq with
@ParseBio
+
@UltimaGenomics
to systematically perturb signaling regulators across six cell lines and learn their downstream targets. Preprint and data at
3
84
358
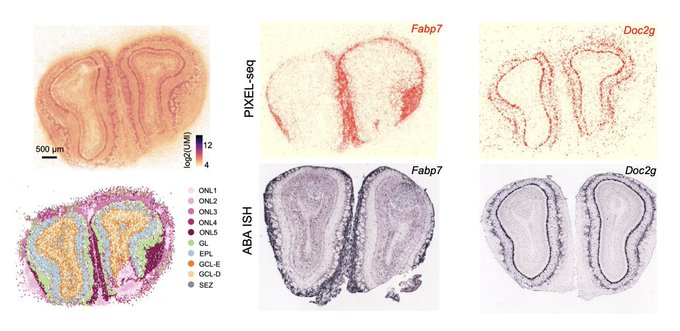
New preprint from
@GuLiangcai
introducing PIXEL-Seq, which creates high-res oligo arrays from polony gels. Performs spatial transcriptomics at 1um resolution with impressive sensitivity:
5
116
353
Single Cell Genomics Day starts tomorrow at 10AM ET!
Come for the awesome keynotes (
@JD_Buenrostro
, Xiaowei Zhuang,
@IdoAmitLab
) and to learn about our favorite new methods and trends for the field in the last year. All talks streamed on Youtube and
5
123
341
Really exciting package from the
@cziscience
#CZCellByGene
team, enabling easy (and free) access to their 33M cell Census dataset in R. We've used it extensively and it works great! Try it out at
Today
#CZCellxGene
is releasing the R package cellxgene.census — it gives access from R to Census, the largest standardized aggregation of single-cell data, composed of 33M+ cells and 60K genes. You can easily export slices to Seurat or
@Bioconductor
. 🧵⬇️
5
191
526
1
88
351
Two really exciting preprints that perform programmable RNA editing (!) using CRISPR-mediated trans-splicing:
* RESPLICE (
@pdhsu
):
* PRECISE (
@jgooten
@omarabudayyeh
):
7
76
348
If you're interested in statistical models of scRNA-seq, this is very clever innovative work from
@const_ae
and
@wolfgangkhuber
. Code is well-documented and open-source, and enabled us to substantially speed up sctransform (update coming soon!):
3
100
347
My overview talk from Single Cell Genomics Day 2024, covering our favorite new technologies and computational methods for the single-cell/spatial field, is now online at
(thanks
@ATCCagan
for the live illustration!)
2
54
344
We're starting a new technology sharing program for
@cegs_ica
!
Sign up for free 'starter kits' for our NTT-seq technology, for simultaneous profiling of multiple chromatin marks in single cells.
8
86
327
Interested in deep learning for scRNA-seq? Check out:
* Autoencoders for de-noising: DCA,
@fabian_theis
;
* Deep transfer learning: SAVER-X, Zhang Lab;
* Single cell variational inference: scVI,
@nir_yosef1
;
2
144
326
New preprint w/
@anshulkundaje
introducing CPA-Perturb-seq! We systematically perturb regulators of cleavage and polyadenylation, and explore post-transcriptional changes at single-cell resolution. Led by
@mh_kowalski
@harm__w
and
@jjohlin
(🧵)
4
87
324
Interested in single cell genomics but need help getting started? Sign up for my lab's free Single Cell Genomics Day on 1/24. Practical talks on new comp. and experimental methods, and keynotes from Aviv Regev, Kun Zhang,
@yimmieg
, and
@roserventotormo
:
13
121
326
Our new preprint, led by
@timoast
and
@aw_butler
. We project CITE-seq data onto HCA references, classify scATAC-seq profiles based on scRNA-seq clusters, and harmonize sequencing and imaging data to predict spatial patterns transcriptome-wide:
5
153
313
Thanks to everyone who tuned in for
#singlecellgenomicsday
! Selected slides, video, reference lists, and resources are now available at
1
87
312
Two papers out today
@NatureBiotech
for jointly analyzing scRNA-seq datasets!
* context-aware batch correction from Marioni Lab
@emblebi
()
* our Seurat ‘alignment’ of experiments across conditions, technologies, and species ()
6
184
314
In our new preprint, led by
@saketkc
, we analyze 58 scRNA-seq datasets - across a wide range of systems, technologies, and sequencing depths - in order to evaluate the choice and parameterization of statistical error models for scRNA-seq:
1
92
311
Andrew Butler (
@aw_butler
), who has been instrumental in the development of our Seurat package and methods for integrative analysis, will be defending his PhD thesis on 4/20! His talk will be online and open to the public- you can register/join at
3
32
303
Thanks to our wonderful speakers, audience members,
@ATJCagan
, and
@genome_gov
(via
@cegs_ica
) for such an enjoyable
#singlecellgenomicsday
2024! After 8 years of SCGD its incredible to see innovation in the field accelerating- see you next year!
5
32
294
Two new methods for integrating single cell data across modalities in
@NatureBiotech
!
1. Mosaic integration (
@shazanfar
/
@MarioniLab
) to 'hop' across datasets:
2. Our bridge integration (
@YUHANHAO2
) implemented in Seurat v5:
1
88
294
Two new preprints for performing differential expression across conditions in scRNA-seq data, *without* having to call clusters first:
1. LEMUR (from
@const_ae
@wolfgangkhuber
):
2. miloDE (from
@Alsu_Troost
@MarioniLab
):
2
92
287
Tremendous advance by
@slinnarsson
lab: EEL-FISH enables multiplexed smFISH of ~500 genes across large tissue volumes. All methods and data are openly released, including spatial atlases of the mouse brain and human cortex:
1
59
291
We've updated our spatial analysis vignette in Seurat to include the recent Slide-Seq2 data from
@macosko
and Fei Chen's lab. The dataset is stunning - after integration with scRNA-seq references, you can precisely localize cell types in the mouse brain:
2
80
284
Our perspective with
@fabian_theis
,
@YUHANHAO2
, and
@mo_lotfollahi
on the power and potential of reference mapping across disease states, molecular modalities, perturbations, and species:
1
74
280
Thanks to everyone who tuned in for
#singlecellgenomicsday
! Selected slides, video, and resources are now available at
For those in a different time zone, we will rebroadcast the entire livestream next Thursday (2/6) at 9:30PM EST.
6
70
276
Thanks to everyone who tuned into
#singlecellgenomicsday
!
We have posted slides, references, and video at
One highlight below -
@JeffreyAFarrell
discusses key practical considerations when analyzing/interpreting a developmental trajectory:
2
95
270
We used our Azimuth human lung reference to map 1M cells (10 studies, >200 donors, 8 disease states) into a common space with harmonized annotations and visualization. You can explore the results using the
@cziscience
cellxgene platform at
1
62
255
Interested in single cell genomics but need help getting started? Check out the agenda for my lab's free workshop, this Friday. Very excited that we will be livestreaming with the support of
@cziscience
! Pls RT and spread the word
4
236
251