Nature Methods
@naturemethods
Followers
148K
Following
2K
Media
2K
Statuses
5K
Nature Methods publishes cutting-edge methods, tools, analyses, resources, reviews, news and commentary, supporting life sciences research. Tweets by editors.
New York
Joined March 2011
Are you curious about using organic dyes in your microscopy experiments but not sure where to start? This Perspective from @rhodamine110 and @jonathangrimm introduces readers to the world of dyes and their pros and cons for biological applications.
7
352
1K
Out today and freely available, a tool many in the image analysis community have been waiting for! . PyImageJ: A library for integrating ImageJ and Python . @EliceiriK @UWMadisonLOCI @ctrueden @CiminiLab @DrAnneCarpenter .
12
338
1K
Out today from the Goodarzi lab! @genophoria SwitchSeeker combines computation and experiment to identify functional RNA structural switches. Applied to the human transcriptome, it identified a novel RNA switch in the 3ʹUTR of RORC, linked to NMD.
0
44
165
It's that time of year!! 🎉We are so excited to announce our 2023 pick for Nature Methods' Method of the Year: methods for modeling development. Read the Editorial to find out why we chose this as our pick. #moty2023.
11
238
652
A Review paper from Peter Kharchenko @KharchenkoLab covers computational methods for scRNA-seq analyses, including their current limitations.
3
248
617
Out today: SCENIC+ is a comprehensive toolbox for inferring and analyzing enhancer-driven gene regulatory networks (eGRN) using single-cell multiomic data. @steinaerts @seppe_winter @Bravo_Potato.
1
204
498
The WashU Epigenome Browser now facilitates integrative visualization and exploration of 1D, 2D and 3D genomic data in a single webpage. #4DN .
2
123
445
A new benchmarking study compares 16 methods for integrating complex single-cell RNA and ATAC datasets and provides a guide to method choice. @MDLuecken @fabian_theis @colomemaria_epi @ICBMunich @HelmholtzMunich.
1
156
440
In a new Comment, Eric Betzig describes his vision for the future of bioimaging and bioimage analysis. He further recommends "Cell Observatories" as a path forward for addressing complex outstanding biological questions. @Eric_Betzig .
0
54
148
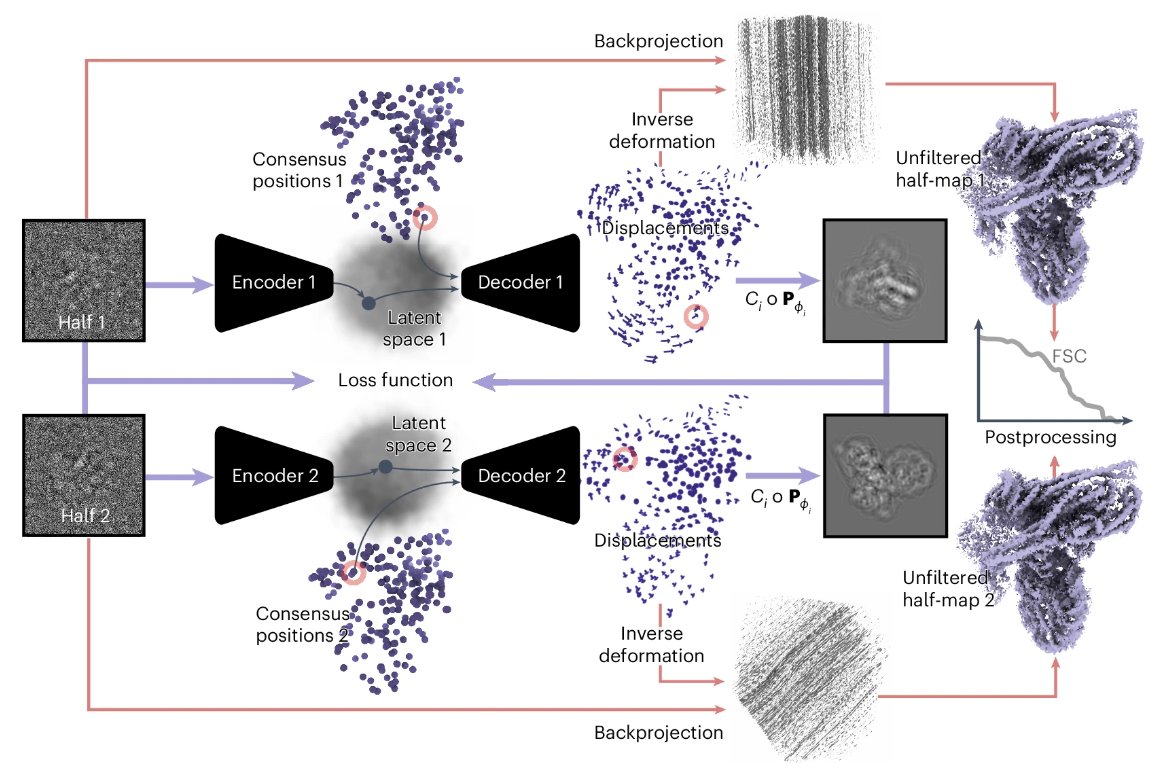
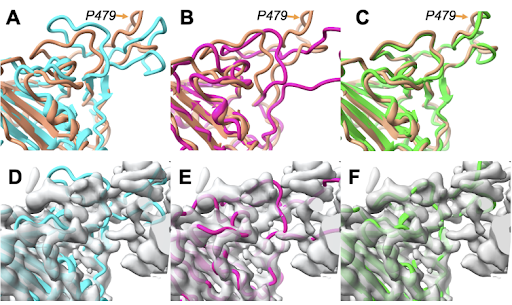
DynaMight from @SjorsScheres @MRC_LMB models continuous structural heterogeneity in cryo-EM datasets, leading to an improved reconstruction of the consensus structure. The study also explores the issue of overfitting when modeling structural flexibility.
0
36
131
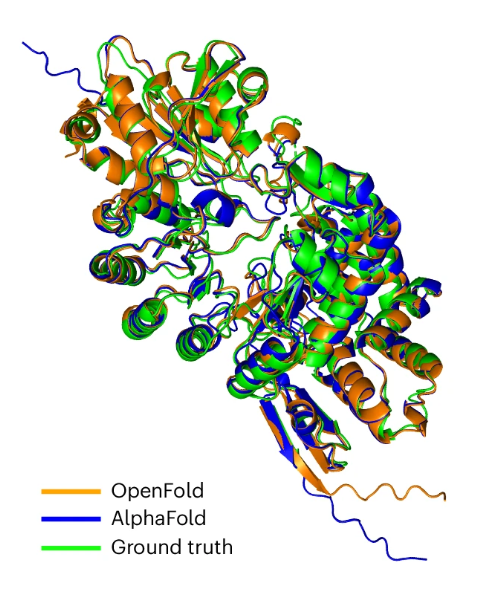
OpenFold developed by @gahdritz, @NazimBouatta, and @MoAlQuraishi is a trainable, open-source implementation of AlphaFold2. It is fast and memory efficient, and the code and training data are available under a permissive license.
6
145
404
Introducing Mitometer, an algorithm for automated segmentation and tracking of mitochondria in live-cell 2D and 3D time-lapse images. @michelledigman @Austin_Lefebvre.
6
100
337
Tangram is a versatile tool for aligning single-cell and single-nucleus RNA-seq data to spatially-resolved transcriptomics data using deep learning. @broadinstitute @tbyanc @gabo_scalia .OA paper:
3
103
320
Squidpy from @fabian_theis and colleagues enables comprehensive analysis and visualization of spatial omics data with high efficiency. OA paper:
0
76
300
Henikoff, @NadiyaKhyzha and @KamiAhmad3 introduce RT&Tag that enables mapping RNA molecules in proximity to a protein of interest in intact nuclei. OA paper:
2
85
286
Bioinformatics advancements unleash the full potential of long-read sequencing in various areas of genomics. A Comment from Michael Schatz (@mike_schatz) and colleagues highlights this thriving domain of active methods development. @JohnsHopkins.
1
85
278
It's finally here! The May issue of @NatureSMB is LIVE and we are excited to share these gorgeous covers with you! 🎉. We split this painting depicting the transport and release of antibody-filled vesicles (from a B-cell) by @dsgoodsell to create these matching covers for #PDB50!
4
54
268
scRNA-seq offers a snapshot of transcriptomes but not RNA dynamics. scNT-seq from the Wu lab @hao_wu_7 integrates metabolic labeling and droplet sequencing to perform parallel analysis of newly-transcribed and pre-existing mRNAs.
5
81
264
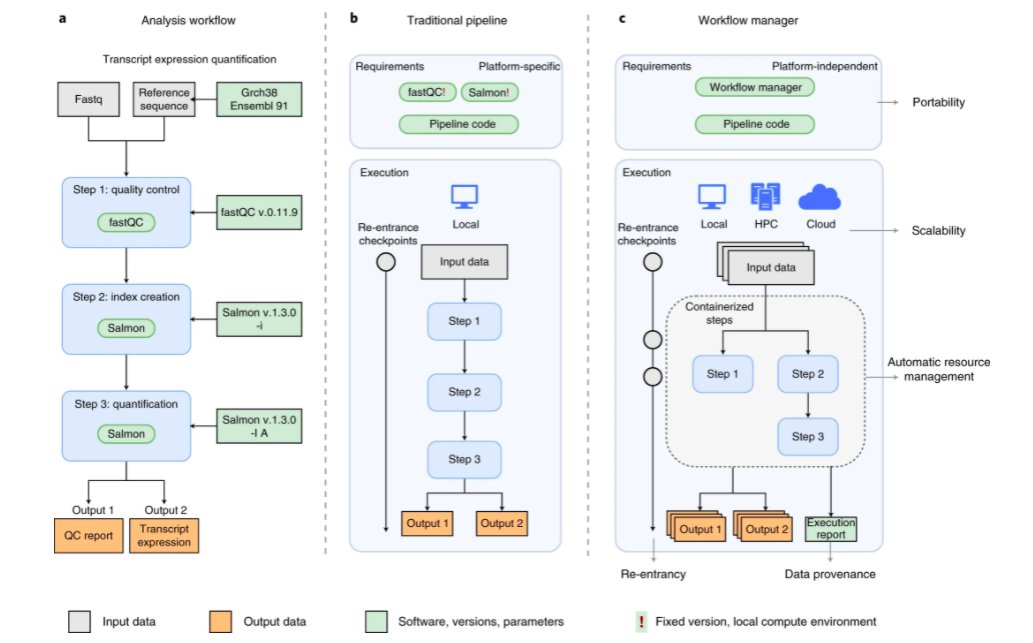
A Perspective highlights workflow managers - useful for developing and managing complex bioinformatics pipelines. .@JonathanGoeke @goekelab @me_myself_andY @astar_gis
1
118
257
Out today, a fantastic Review on spatial omics and multiplexed imaging methods for exploring cancer biology from Shalin Naik and Kelly Rogers @shalinhnaik @KRogers1001
2
92
262
This Review from @OliverStegle and @BrittaVelten discusses statistical and computational strategies for analyzing various spatial and temporal omics data types, with an emphasis on the common modeling principles.
0
80
253
The Meltome Atlas compiles the thermal stability of 48,000 proteins across 13 species ranging from archaea to humans, providing a resource for analyzing protein stability in the context of function and interactions. @kusterlab @savitski_lab
6
141
254
In their Comment, AlphaFold developers John Jumper and @demishassabis of @DeepMind describe the design of AlphaFold’s neural network and give their views on how it will transform the field of structural biology.
0
54
255
Out yesterday from @adam_k_glaser and @jonliu123, a versatile hybrid open top light sheet microscope for imaging of large cleared samples. Check out their images! 🤩
2
50
254
Is cell segmentation a solved problem? Maybe not yet, but the results of this multimodality cell segmentation challenge get us closer! (Spoiler, the winner is a generalist Transformer-based model). @BoWang87
1
52
257
Come see CellPose from the Pachitariu lab for your image segmentation needs! Cellpose is a generalist, deep learning-based approach for segmenting structures in a wide range of image types. @marius10p
1
63
243
Our March issue is live! 🎉On the cover, multiplexed tissue imaging evokes a stained glass window. This gorgeous image was created by Stefan Uderhardt @red_ven and Andrea Radtke.
4
50
209
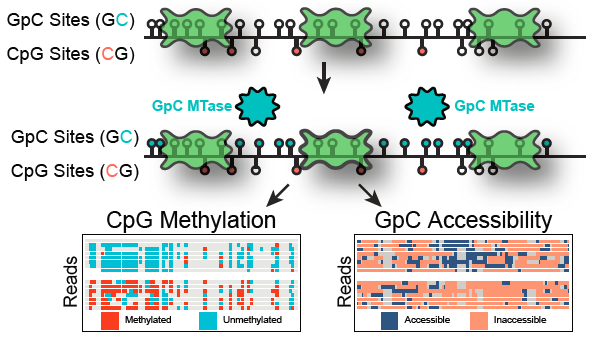
Nanopore sequencing allows profiling long-range patterns on single molecules. nanoNOMe uses nanopore sequencing to profile CpG methylation and chromatin accessibility in human cells. @timp0 .
2
59
210
An iterative approach for improving #AlphaFold protein model quality using experimental density maps, where rebuilt models are used as templates in new AlphaFold predictions, from @TerwillTom and colleagues. OA paper:
3
70
204
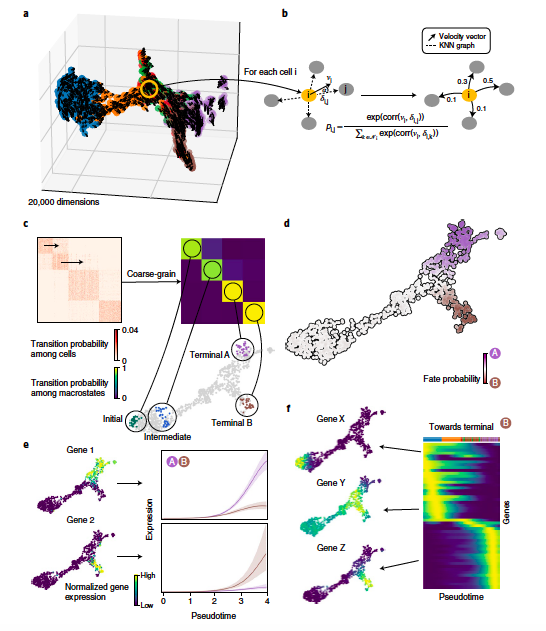
CellRank infers directed cell state transitions and cell fates using RNA velocity information from single-cell data. @MariusLange8, @dana_peer, @fabian_theis, @sloan_kettering, @ICBmunich.OA paper:
0
66
204
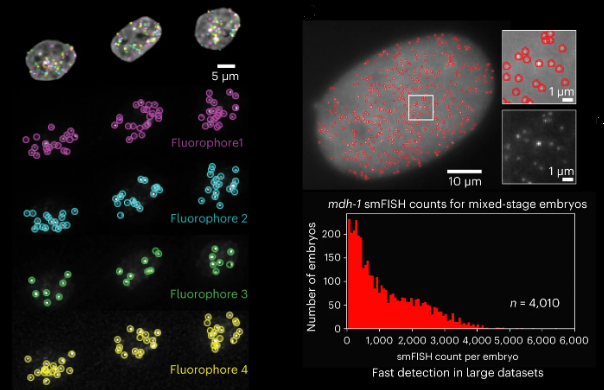
Out today! Radial Symmetry-FISH (RS-FISH) -- an accurate, fast, and user-friendly software for spot detection in 2D and 3D images. Bring on your smFISH, spatial transcriptomics, or spatial genomics applications! @kisharrington @successprocess @preibischs
2
50
199
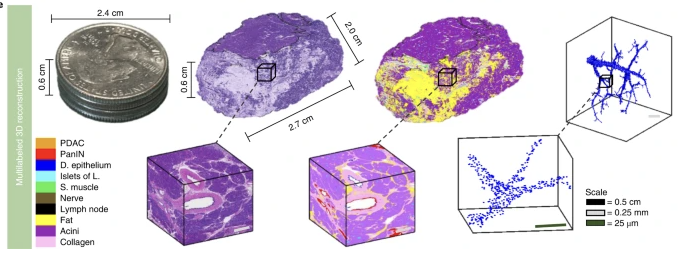
CODA is a method for 3D reconstruction of large tissues at subcellular resolution from serially sectioned H&E images, from Pei-Hsun Wu, @deniswirtz and colleagues. @johnshopkins @ashleykiemen.
0
65
198
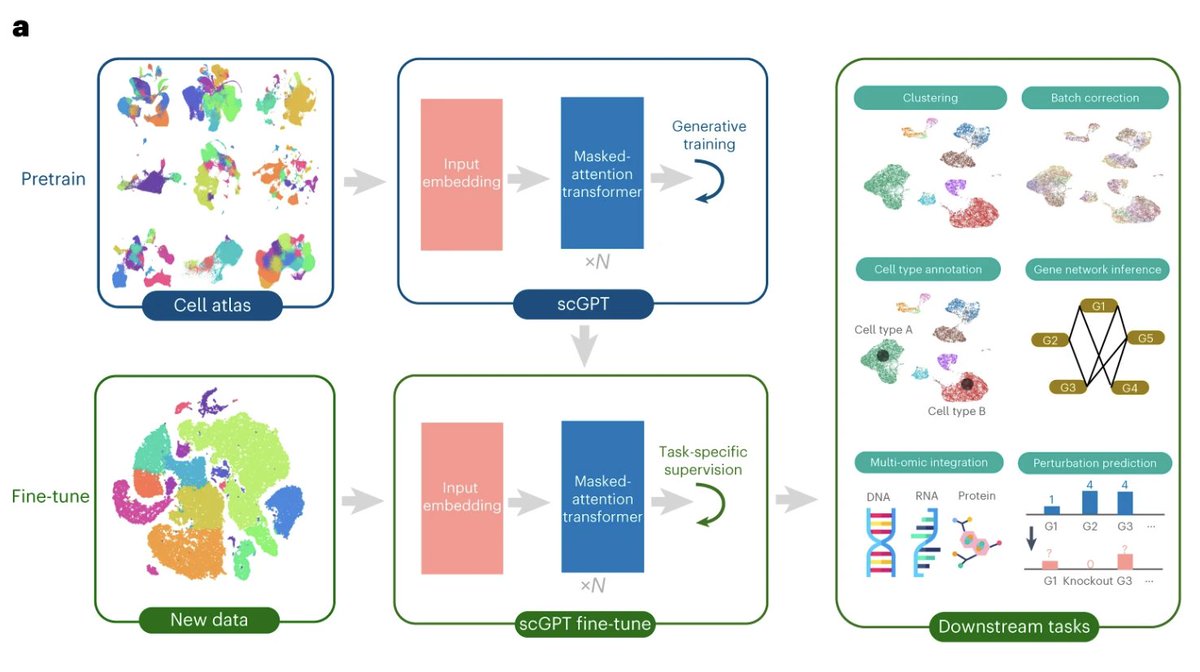
Pre-trained using over 33 million single cell RNA-seq profiles, scGPT is a foundation model facilitating a broad spectrum of downstream single-cell analysis tasks by transfer learning. @BoWang87 @HAOTIANCUI1 @ChloeXWang1 @UofT @VectorInst @UHN .
3
52
204
Out today from @bewersdorflab! Two-color fluorogenic DNA-PAINT introduces self-quenching, kinetics-optimized probe designs. This approach improves imaging speed 26-fold and eliminates the need for optical sectioning.
3
59
193
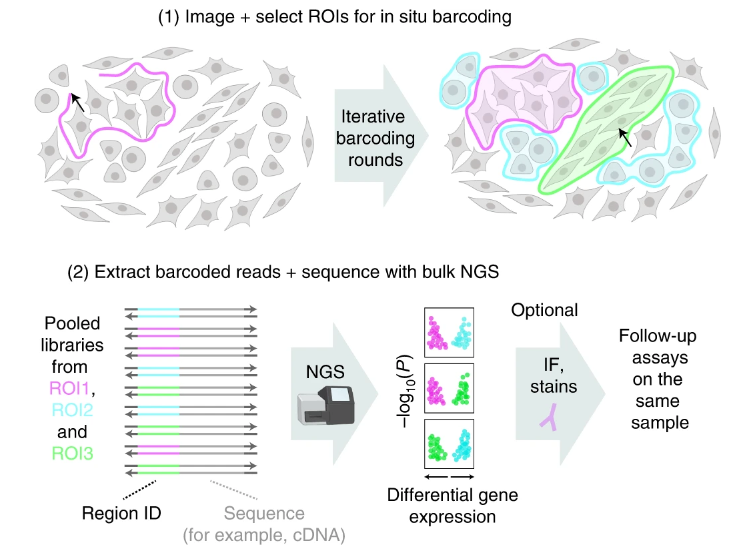
What's new in spatial 'omics? Light-seq from the @SinemKSaka1, Kishi, Cepko, and Yin labs! Light-Seq uses light-directed DNA barcoding in fixed cells and tissues for multiplexed spatial indexing and subsequent next gen sequencing of even rare cell types.
1
46
194
GlycoRNA is a thing!! 😮. (this eloquent tweet by @rita_strack, twitter takeover).
Excited to share my work w/@CarolynBertozzi describing the discovery of glycoRNA today @CellCellPress. Since our @biorxivpreprint we uncovered an unexpected twist: glycoRNAs are on the surface of cells+some Siglec receptors have RNA-dependent cell binding
3
33
193
Our February issue is live! 🎉 On the cover, a beautiful example of cryo-expansion microscopy (Cryo-ExM) used to observe a human cell in mitosis, from @MarineLaporte8 of @CentrioleLab. OA paper here:
1
40
192
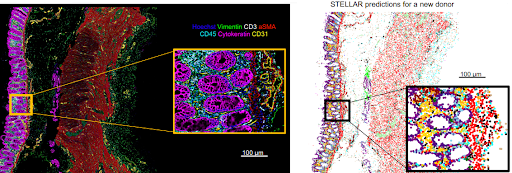
Introducing STELLAR: a geometric deep learning method for cell type discovery and identification in spatially resolved single-cell datasets. @jure @GarryPNolan @SnyderShot
0
51
193
The PanGenome Research Tool Kit (PGR-TK) achieves flexible and scalable representation, visualization, and analysis of genomic variation using pangenome graphs. @infoecho @sedlazeck @GenomeInABottle @psudmant @srbehera11 .
1
76
185
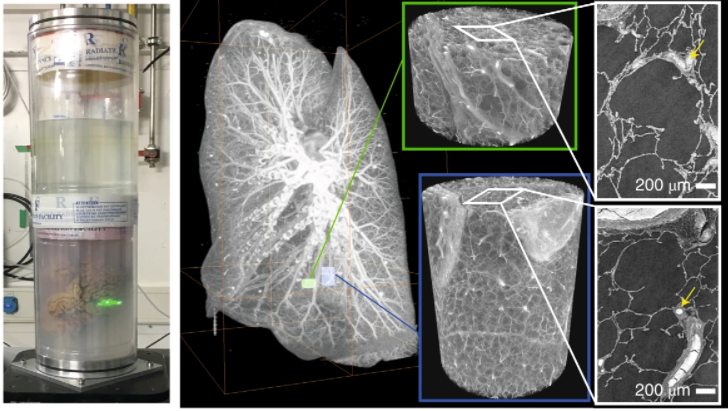
Our May issue is now live! 🎉.On the cover, a beautiful optically cleared mouse brain imaged with a hybrid open-top light-sheet microscope, from @adam_k_glaser (paper here: .
0
39
178
The Signac framework enables end-to-end analysis of single-cell chromatin data and offers interoperability with the Seurat package for multimodal analysis. @timoast @satijalab @nygenome
0
59
183
Extra-long tandem repeats (ETRs) such as centromeres and immunoglobulin loci are difficult to align. UniAligner is a computational method for efficiently and accurately aligning ETRs, facilitating analysis of their variation and evolution. @AndreyBzikadze.
1
58
179
Really pleased to publish this timely set of checklists to help improve the presentation and reproducibility of microscopy data. Wonderful community effort led by @ChriSchmied @helenajambor @tischitischer
1
74
181
A new paper presents a statistical framework for power analysis of spatial omics studies, facilitated by an in silico tissue generation method. @ethanagb @DenisSchapiro @mssanjavickovic @broadinstitute @HiTSatHarvard . OA paper:
4
58
179
Using transcriptomic similarity and spatial coordinates, PASTE allows aligning and integrating spatial transcriptomics data generated from adjacent tissue slices. @RonZeira @benjraphael.
1
54
168
An automated multi-scale 3D imaging platform enables both organoid culture and high-content imaging on a single instrument. @Viasnofflab @BeghinTeam
1
52
170
Research Highlight by @DrArunimaSingh:.@cees_dekker and colleagues @tudelft have expanded nanopore-based single-molecule sequencing technology to post-translational modifications. They demonstrate the detection of phosphorylation along single peptides.
1
54
177
Out today from the @davidvanvalen lab! DeepCell Kiosk: cloud-native software that dynamically scales deep learning workflows to accommodate large imaging datasets.
1
36
174