Bo Wang
@BoWang87
Followers
10K
Following
51K
Media
140
Statuses
1K
Assistant Prof @UofT; CIFAR AI Chair @VectorInst; Chief AI Officer @UHN; former PHD, CS @Stanford; opinions my own. #AI #healthcare #biology #LLM
Toronto, Ontario
Joined September 2016
(1/5) Attention, Cellular Imaging Enthusiasts! . Thrilled to share that our Analysis "The Multi-modality Cell Segmentation Challenge: Towards Universal Solutions" has been published in Nature Methods (@NatureMethods) 🎉🎉🎉 . Cell segmentation is a critical component of
29
282
1K
Most MIT profs who I know are extremely honest and morally upright. I hope this is an exception.
Mitigating racial bias from LLMs is a lot easier than removing it from humans! . Can’t believe this happened at the best AI conference @NeurIPSConf . We have ethical reviews for authors, but missed it for invited speakers? 😡
16
46
937
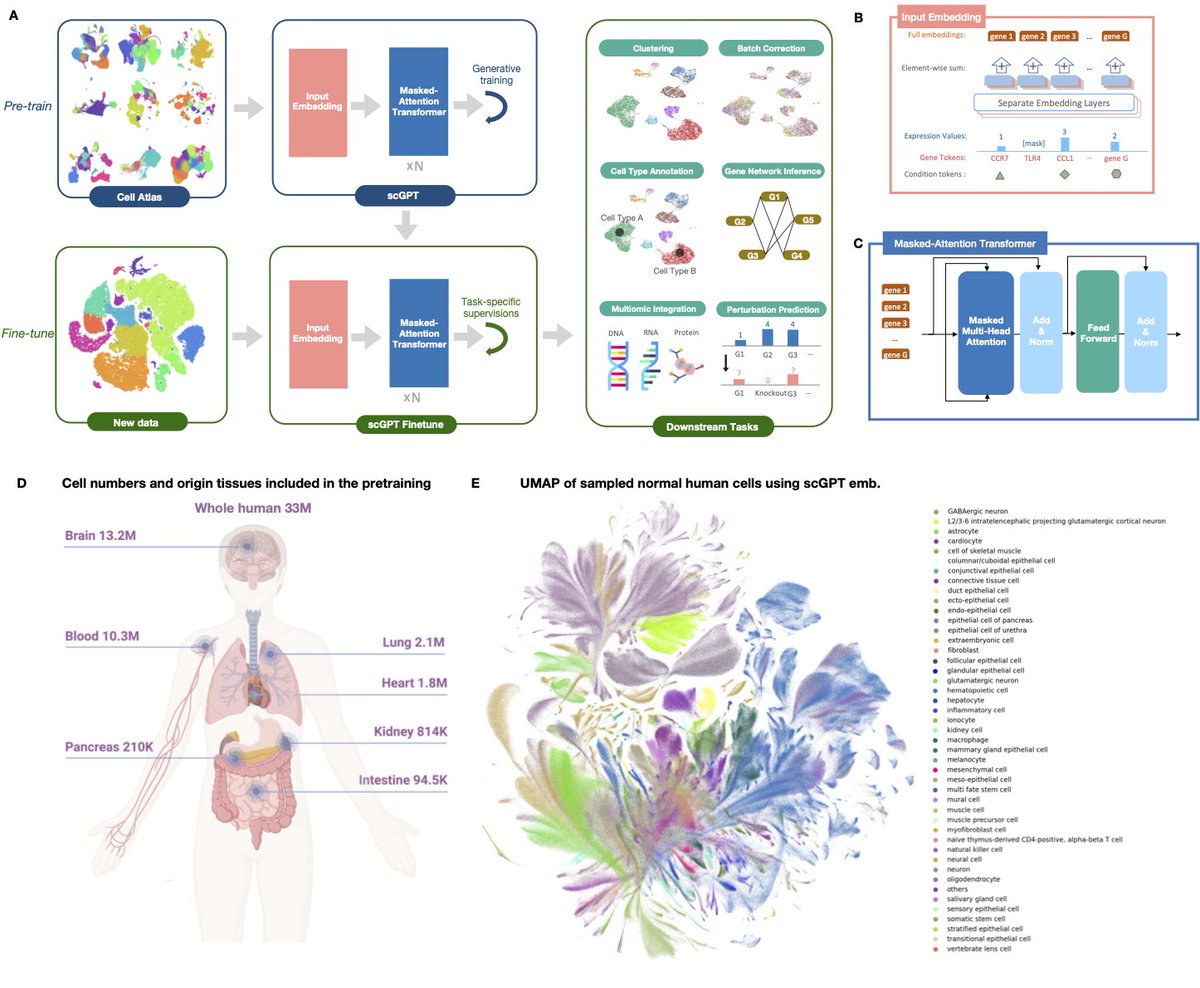
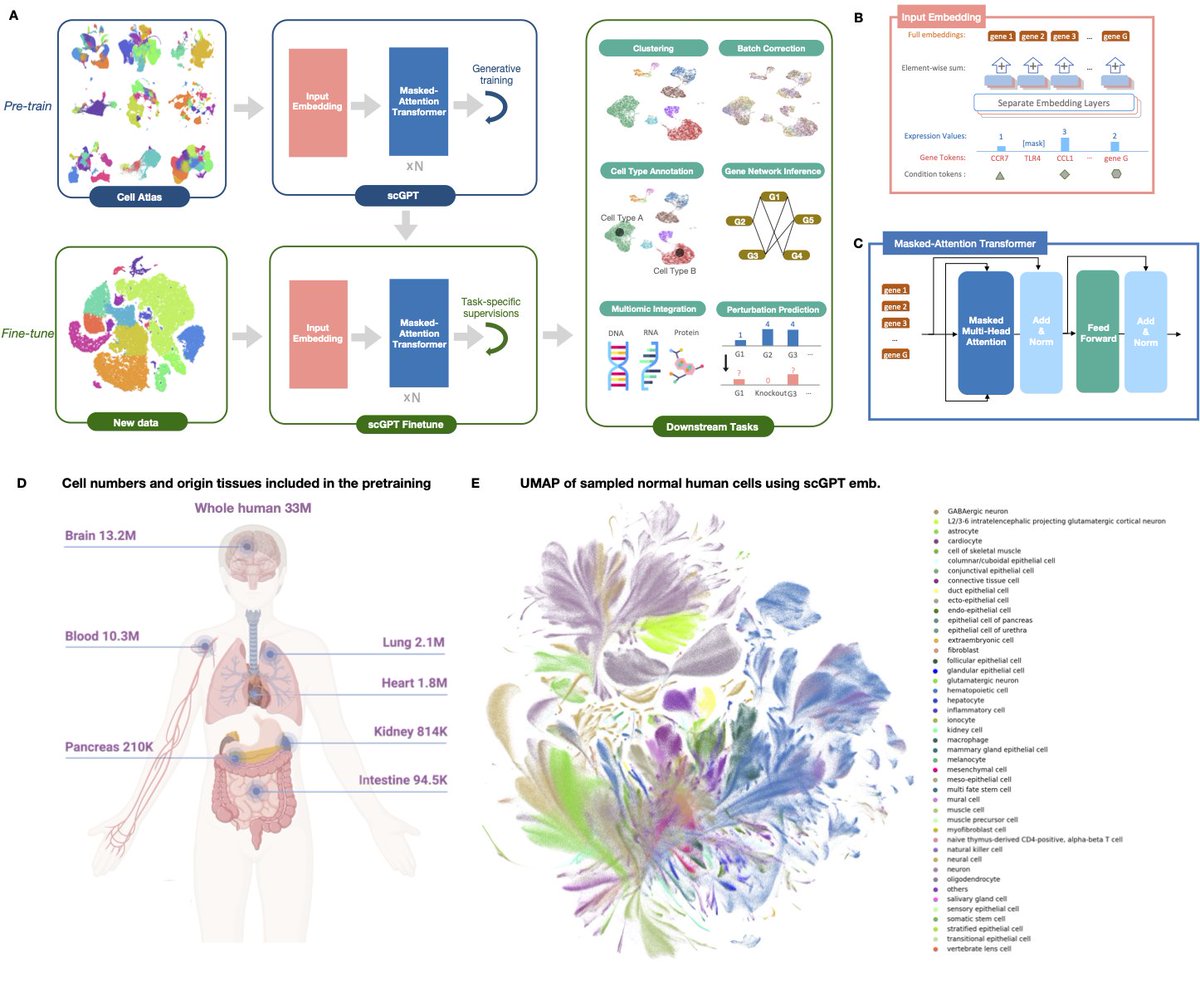
🔬 Exciting News! Our manuscript, "scGPT: toward building a foundation model for single-cell multi-omics using generative AI" is now finally published in Nature Methods (@NatureMethods) 🎉 !!! . (Re-)Introducing scGPT: A transformative foundation model engineered for single-cell
43
231
907
Truly humbled to be promoted as the Chief AI Scientist at University Health Network, Canada's largest research hospital. My deepest gratitude to everyone who has supported and believed in me, especially the dedicated trainees and students in the lab. As we look to the future, I'm.
Congratulations to Dr. @bowang87 UHN's Chief Artificial Intelligence (AI) Scientist. @camillebains1 of Canadian Press outlines how the pioneering role builds on the @UHNAIHUB and its potential to use machine learning to improve health care. Read more →
67
32
485
couldn’t agree more about the need for more computational biologists. More importantly, we must ensure they receive the recognition and support they deserve. Their work should never be dismissed as merely “just service”. Additionally, I’d like to remind everyone of the excellent
WE NEED COMPUTATIONAL PEOPLE IN BIOLOGY. WE NEED THEM IN DRUG DISCOVERY AND MULTI OMICS. WE NEED HARDWARE PEOPLE TO BUILD SPECIFIC AND PORTABLE DEVICES. BUT MOST IMPORTANTLY WE NEED DEVS. WE NEED TO MAKE OPEN ACCESS DATA AS ACCESSIBLE AS POSSIBLE. WE NEED COMPUTE GRANTS. GIVE.
12
127
483
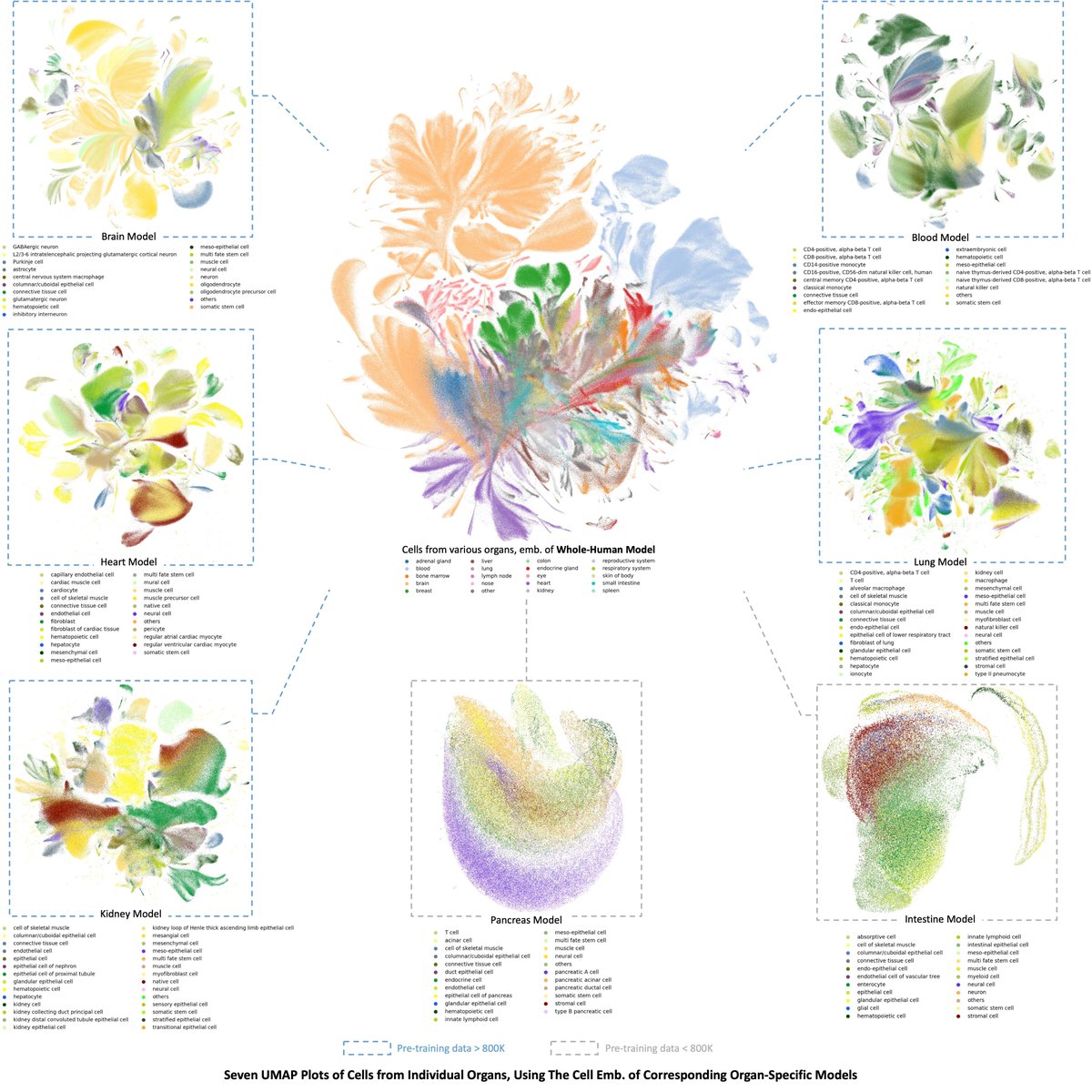
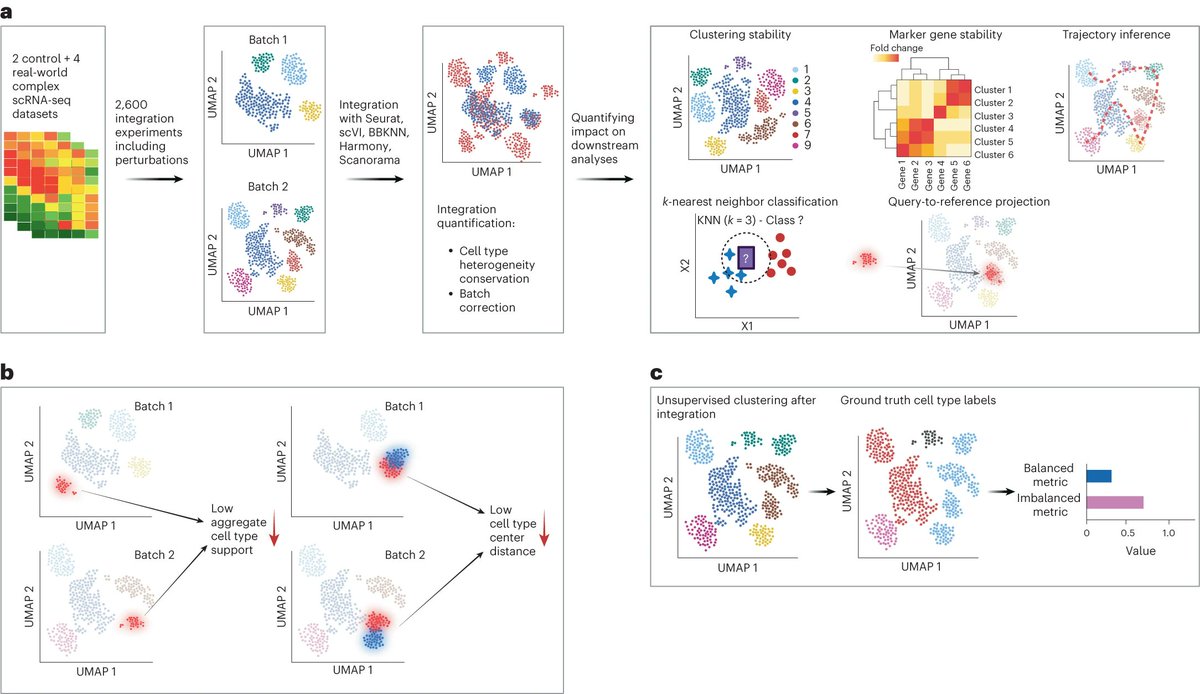
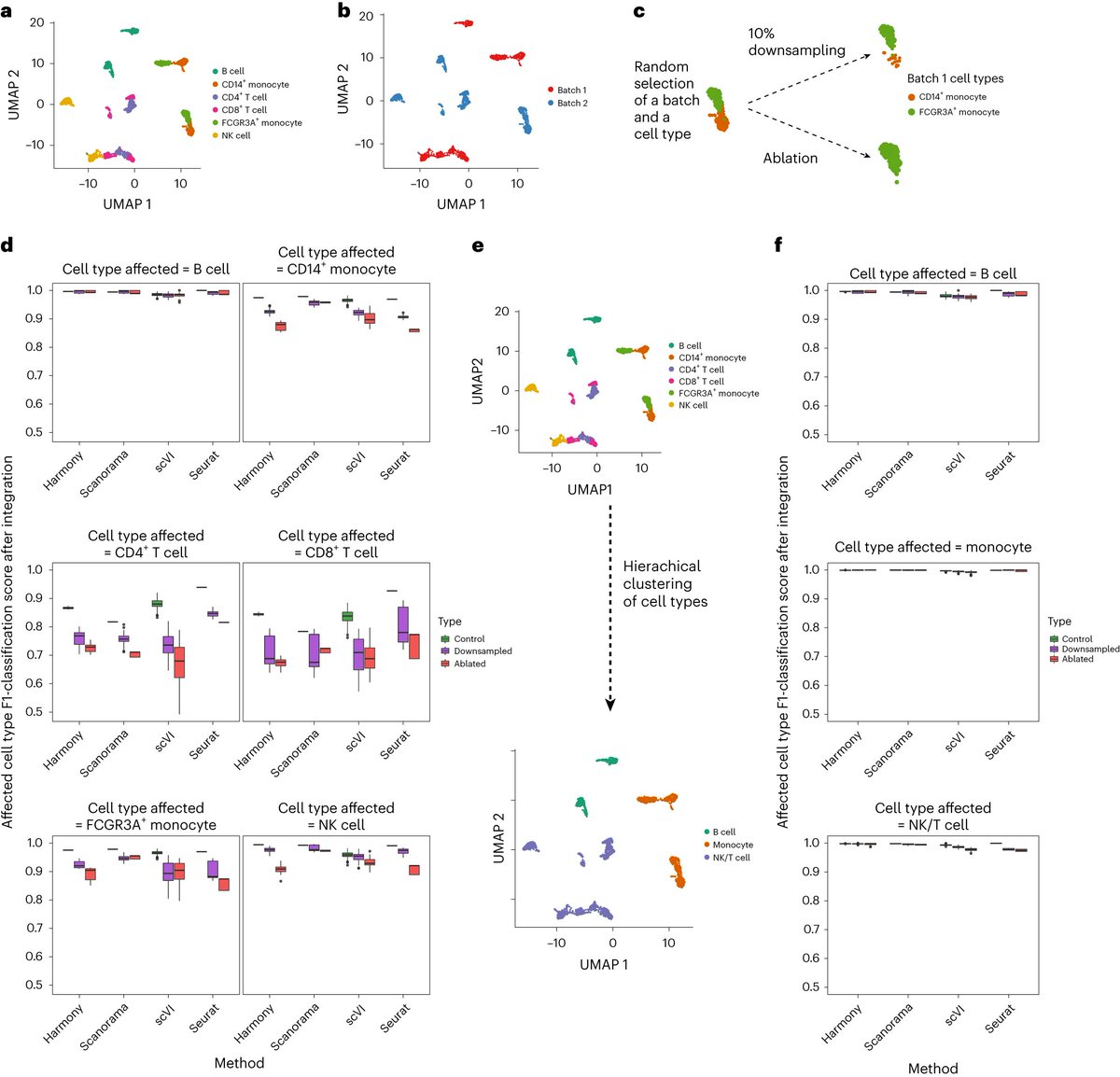
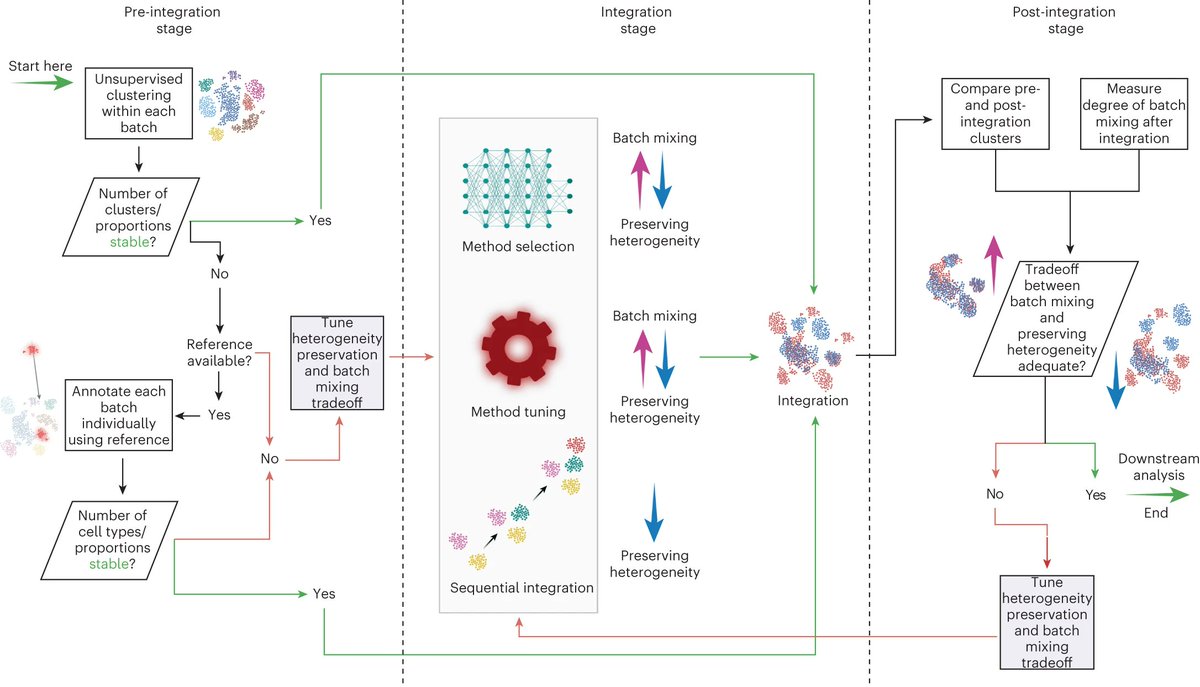
**Is single-cell data integration a solved problem? **. Lots of great methods, but still not quite yet! Specifically, Check Dataset Imbalance! . 🎉 Excited to announce that our study on dataset imbalances in single-cell analysis just hit Nature Biotechnology (@NatureBiotech)! 🎉
15
89
426
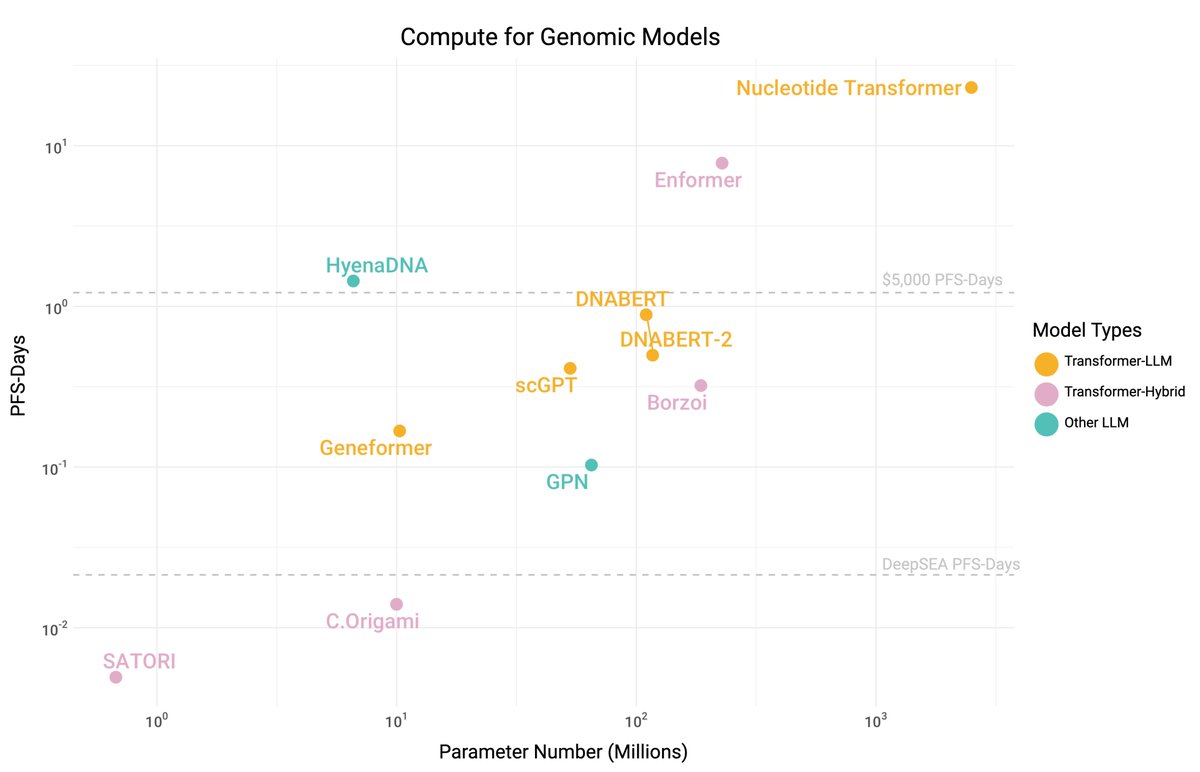
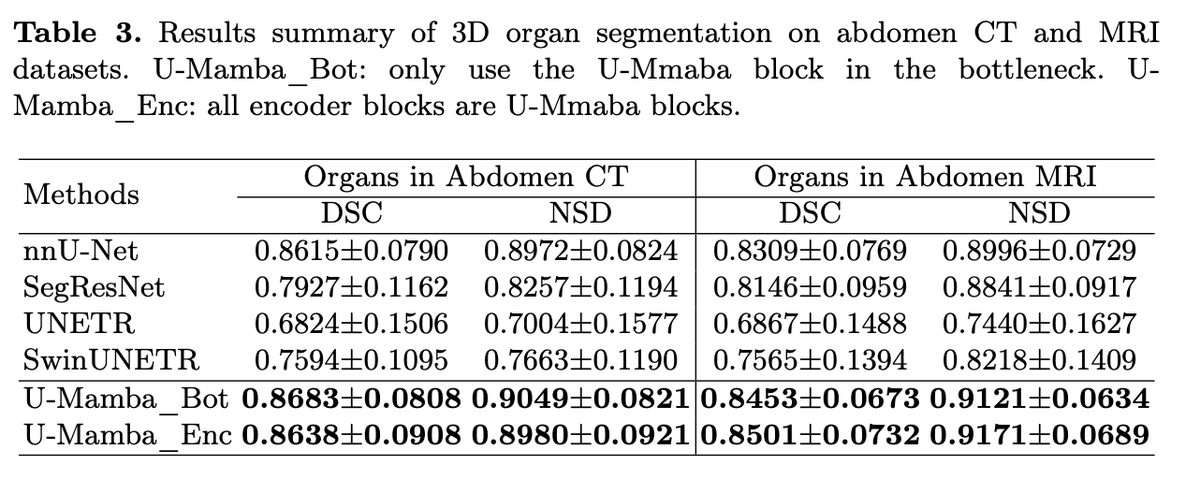
Our recent preprint U-Mamba ( has sparked lots of interest in State Space Models (SSMs) or the Mamba model, particularly its adoptions in vision. Many people have reached out to request more educational materials for this new alternative to Transformers.
🎉 Thrilled to announce the very first 2024 preprint from our lab: U-Mamba 🐍, a novel backbone for universal bio-medical image segmentation!. Background: Transformers have not transformed bio-medical images. While Transformers excel in general vision and language tasks, they
6
81
397
(Re-)Introducing #ClinicalCamel 🐪: an innovative open-source conversational #LLM engineered specifically for healthcare: . Paper: Model weights in HF: Eval Scripts at Github: Live Demo:
10
104
368
One of the biggest challenges in #AI and #healthcare is how to collaboratively train large AI models from multiple hospitals without compromising patient privacy? . Thrilled to introduce our new paper, DeCaPH, which is now officially published in @TheLancet @eBioMedicine!
11
88
329
Thrilled to present our latest commentary in #NatureMethods on foundation models for biological image segmentation! A central task of leveraging biological images is the extraction of quantitative features that can be used to characterize and compare.
5
53
275
@drkeithsiau Our medical chatbot (named Clinical camel, ) says it is Peutz-Jeghers Syndrome (PJS).
6
33
264
After labor-intensive debugging, we finally deployed our MedSAM2 models available on Hugginface for medical and biological video segmentation. Welcome to try it. There are three advantages of this API: and upload videos of any length, no limitation on
🚀 The Segment Anything Model (SAM) has been upgraded to SAM2, featuring an efficient image encoder for segmenting images and videos. But does SAM2 outperform SAM1 in medical image and video segmentation?. We're thrilled to present our paper "Segment Anything in Medical Images
5
64
263
🚀🌱 Large language of life models: foundation models for longevity and aging! . Our lab has recently been involved in two groundbreaking DNA methylation foundation models: CpGPT and MethylGPT! These “Large Language of Life” models (@EricTopol) mark a new era in aging.
11
57
216
I had the privilege of meeting Demis Hassabis and John Jumper at the Gairdner dinner in Toronto last year, where we discussed what the “next AlphaFold moment” might look like. During our conversation, Demis shared his long-standing dream of creating virtual cells—an incredibly.
Winning the @NobelPrize is the honour of a lifetime and the realisation of a lifelong dream - it still hasn’t really sunk in yet. With AlphaFold2 we cracked the 50-year grand challenge of protein structure prediction: predicting the 3D structure of a protein purely from its.
6
30
257
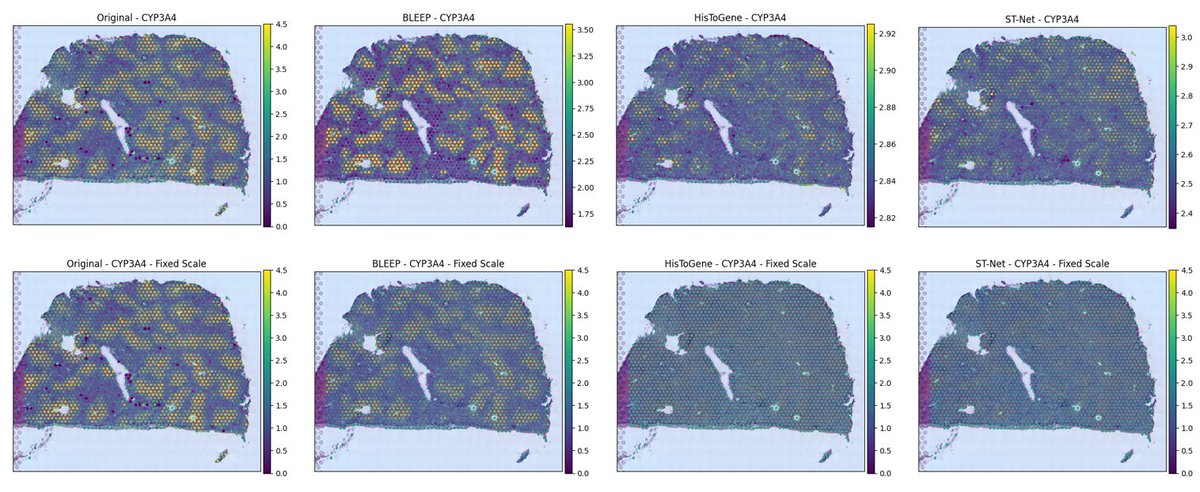
🔬Can whole-slide Hematoxylin and eosin (H&E) stained histology images predict gene expression profiles in single-cell spatial omics? #spatial #SingleCell. Introduce our latest paper, BLEEP (Bi-modaL Embedding for Expression Prediction): a state-of-the-art tool predicting spatial
2
69
250
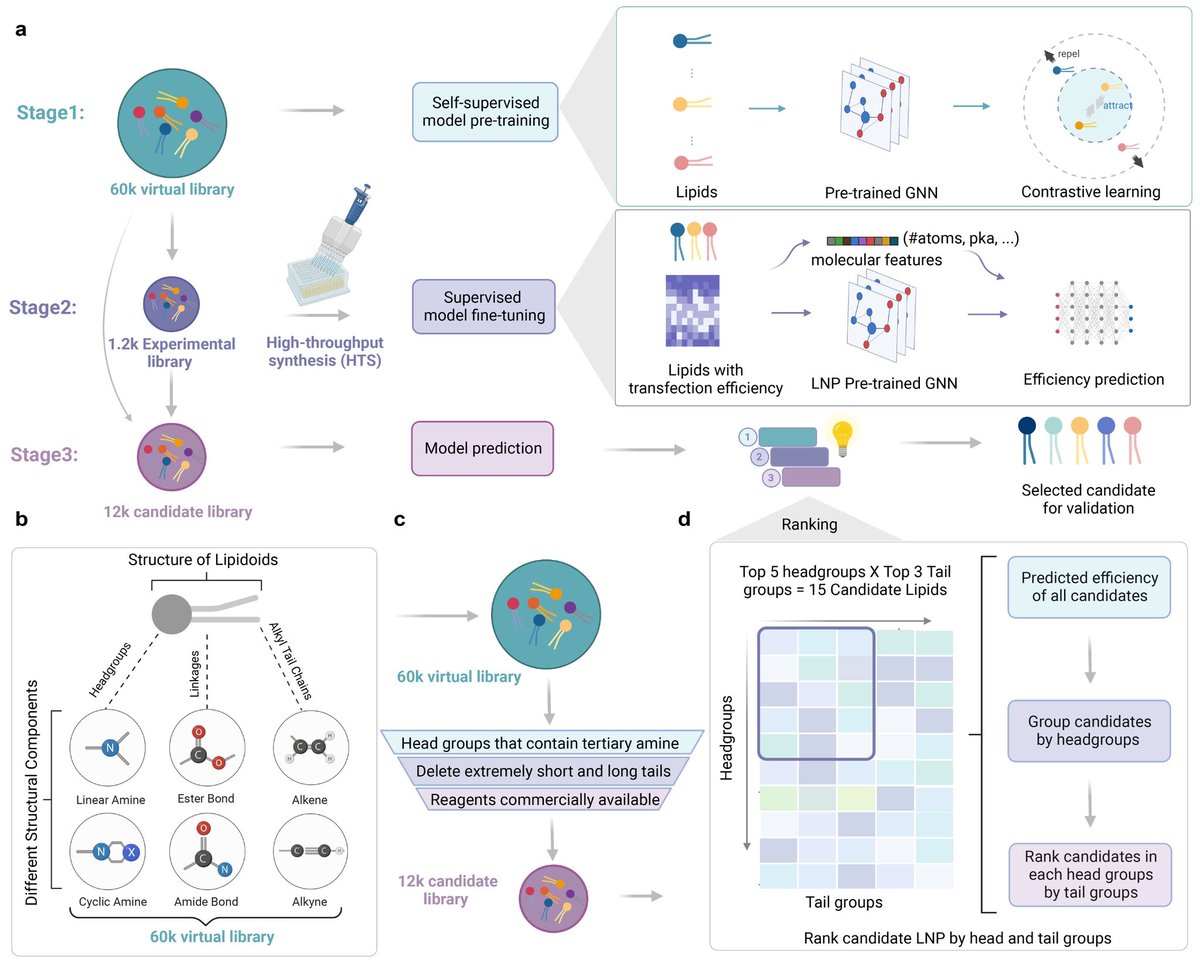
mRNA medicine has changed the world. Wonder how #AI can help with mRNA therapy development? Thrilled to share our latest collaboration with @BowenLi_lab! Our paper, "AGILE Platform: A Deep Learning-Powered Approach to Accelerate LNP Development for mRNA Delivery," showcases how
5
82
242
Another amazing breakthrough of Mamba in Vision is just out: Vision Mamba (Vim) (paper: , a great work from @XinggangWang's group! . What is Vision Mamba?. Vision Mamba (Vim) is not just another model; Departing from conventional attention mechanisms, Vim
4
46
238
🚀 The era of foundation models for biology and medicine is here—and it’s on fire! 🔥. Our lab is at the forefront of this revolution, building cutting-edge models to transform biomedical research and healthcare:. 🔬 scGPT: Redefining single-cell omics with advanced foundation.
During my sabbatical, one project I really enjoyed was training DNA foundation models with the team at @arcinstitute. I've long believed that deep learning should unlock unprecedented improvements in medicine and healthcare — not just for humans but for animals too. I think this
2
39
233
✨ Excited to announce the first Workshop on Foundation Models for Medical Vision in #CVPR2024! . 💡Explore the potential of foundation models in medical imaging to augment human intelligence and revolutionize patient care. ❤️🔥We are proud to host an esteemed lineup of speakers:
3
53
209
Thrilled to announce that our AGILE framework, an AI-guided LNP development for mRNA delivery, is now published in @NatureComms! 🎉 This is a collaborative work with @BowenLi_lab. (Re-)Introducing the AGILE Platform: A Deep Learning-Powered Approach to Accelerate LNP Development
mRNA medicine has changed the world. Wonder how #AI can help with mRNA therapy development? Thrilled to share our latest collaboration with @BowenLi_lab! Our paper, "AGILE Platform: A Deep Learning-Powered Approach to Accelerate LNP Development for mRNA Delivery," showcases how
2
43
187
Great News: DeepVelo finally published in Genome Biology (@GenomeBiology)! 🎉 🙏❤️🔥. DeepVelo: Deep learning extends RNA velocity to multi-lineage systems with cell-specific kinetics. In-press: . #bioRxiv preprint (April 2022) :
4
28
174
This is incredible! This trend, once unique to computer science, is now emerging in life sciences too. As AI continues to evolve, it prompts a big question: What does the future hold for academia? Could we see a shift, or even a disappearance, of traditional academic roles?.
One of the biggest cultural shifts in life science PhD programs over the past decade - industry directly replacing academic jobs for graduates. Source: @NSF @DrChrisTSmith
21
23
171
Absolutely! I’ve seen it happen three times in my lab in the last year: We post our papers as preprints and share our codes on GitHub early, but then others slightly modify our work and get published first. Meanwhile, our papers languish in long review processes. To add insult to.
@J_my_sci It depends. I personally prefer to preprint but we have noticed some labs repeating our experiments by monitoring our preprints. This is starting to happen to other labs as well as publishing can take months to years after submission while other journals it can take days.
17
23
167
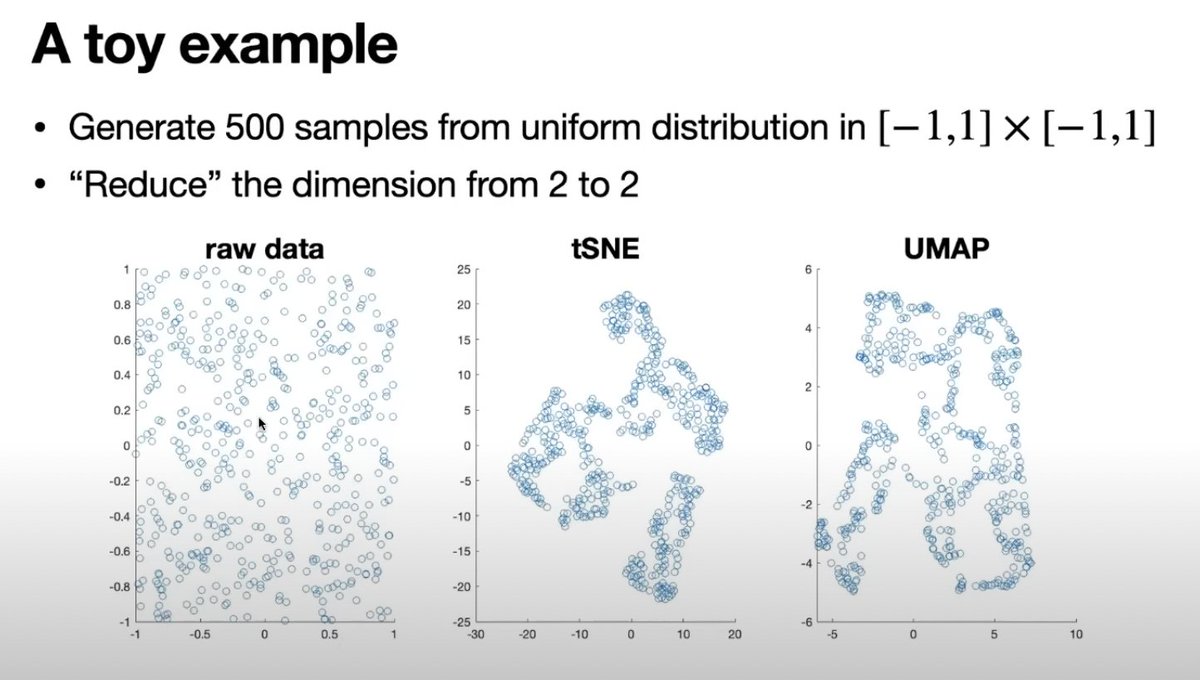
It's clear we need a visualization method for genomic data analysis that prioritizes reliability and honesty over aesthetic appeal. While tSNE and UMAP may yield visually pleasing results, a more truthful representation of the data's distribution should be the focus.
This toy example may change how you look at tSNE & UMAP. They find clusters in the most uniform data possible.
12
28
151
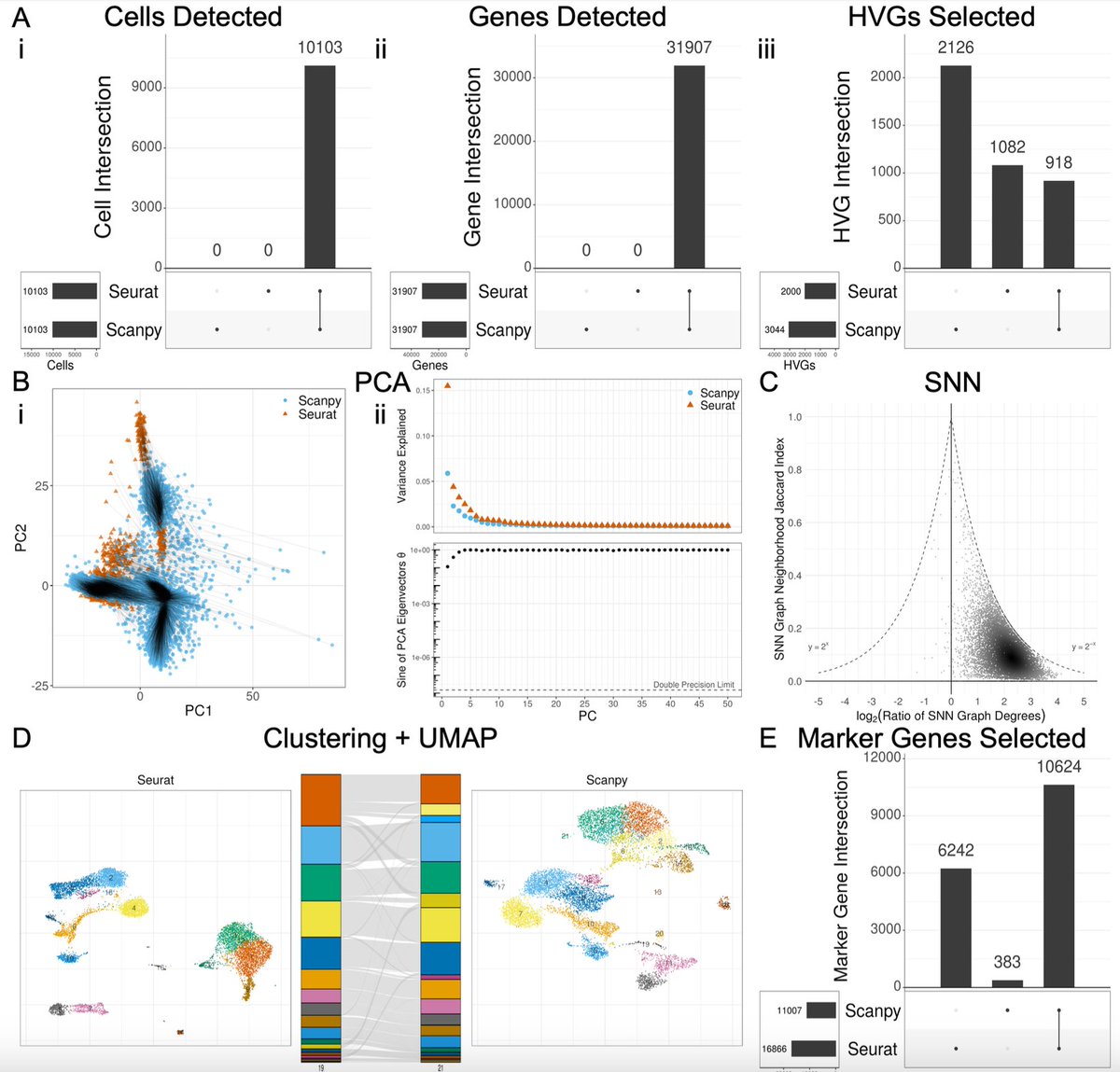
This study reveals something fascinating: Different packages (e.g., in terms of versions and programming languages) of the same algorithms/pipelines can lead to varied results in downstream analysis. Here are my thoughts:. Firstly, this should not be used to critique Seurat and.
The choice of whether to use Seurat or Scanpy for single-cell RNA-seq analysis typically comes down to a preference of R vs. Python. But do they produce the same results? In w/ @Josephmrich et al. we take a close look. The results are 👀 1/🧵
4
32
149
Summary of 2023:.______________________________________________________. Without any doubt, 2023 is the year of LLMs/Generative AI. As a researcher in #AIforHealth and #AIforBiology, 2023 has been a GREAT year for me and the lab! Today is the perfect moment to reflect on our
9
21
146
Our BLEEP is accepted by #NeurIPS2023 ! Congratulations to the team @RonaldXie1 @garybader1 ! BLEEP is ground-breaking because it finds predictive powers from the H&E images to gene expressions in spatial single-cell data! @VectorInst @UofTCompSci @UofT_LMP @DonnellyCentre.
🔬Can whole-slide Hematoxylin and eosin (H&E) stained histology images predict gene expression profiles in single-cell spatial omics? #spatial #SingleCell. Introduce our latest paper, BLEEP (Bi-modaL Embedding for Expression Prediction): a state-of-the-art tool predicting spatial
7
22
153
Thanks @naturemethods for highlighting our work! Cell segmentation is moving forward fast @rita_strack 🔥🔥🙌🙌.
Is cell segmentation a solved problem? Maybe not yet, but the results of this multimodality cell segmentation challenge get us closer! (Spoiler, the winner is a generalist Transformer-based model). @BoWang87
4
19
145
(1/4) Our BIONIC is finally published in @naturemethods (! Huge congrats to Duncan who really pulled this work off! Great teamwork with my amazing colleagues @garybader1 and Charlie Boone! @PMunkCardiacCtr @VectorInst @UofT_TCAIREM @LMP_UofT @UofTCompSci.
5
47
142
Exciting news! Our MedSAM is featured in Computer Vision News! 🎉🎉🎉🙌. Check it out: We are not stopping at papers and aiming to take integration to the next level by incorporating MedSAM with 3D Slicer, a top-tier medical imaging viewer. 🏥. **Segment
🎉 The best way to start the week is to find out that our MedSAM is finally published today in Nature Communications!. **Segment anything in medical images** .Paper: arXiv: Data & Code: MedSAM is the first
8
38
145
Due to numerous requests, I'm providing unrestricted (no paywall), view-only access to the paper through this SharedIt link: Appreciate the support!. PS: to avoid the slightest possibility of confusion, scGPT is unrelated to UMAP; it is not a.
🔬 Exciting News! Our manuscript, "scGPT: toward building a foundation model for single-cell multi-omics using generative AI" is now finally published in Nature Methods (@NatureMethods) 🎉 !!! . (Re-)Introducing scGPT: A transformative foundation model engineered for single-cell
7
26
142
Very honored to be awarded with Gairdner Early Career Investigator Award! #AI is really changing the landscape of biology, particularly single-cell analysis! Thanks for the recognition from @GairdnerAwards! Excited to meet with this year's Gairdner Laureates and discuss science!.
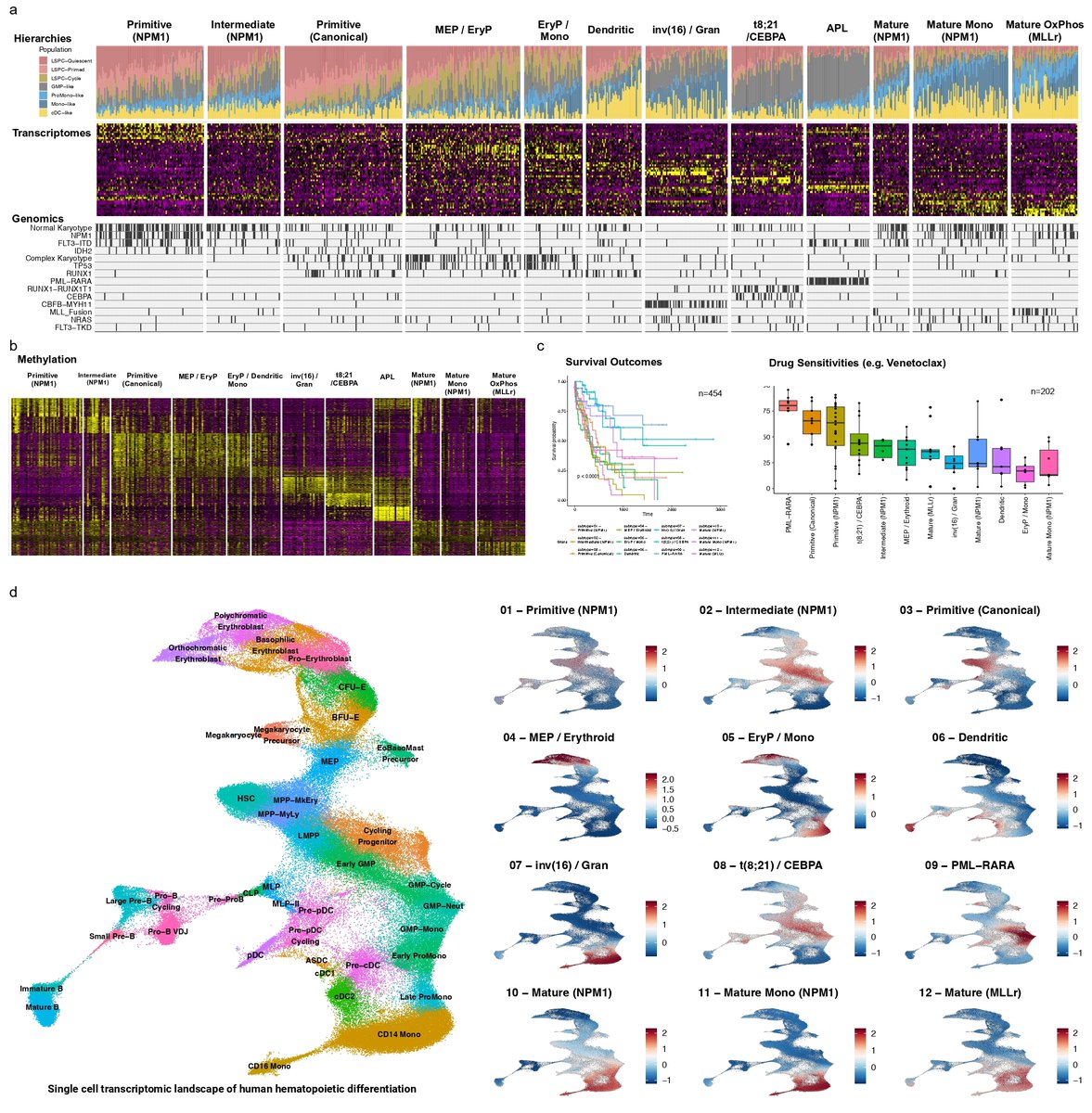
Dr. Bo Wang (@BoWang87) from @VectorInst was one of two winners selected by Drs. John Dick and Stuart Orkin. Dr. Wang was selected for his work on machine learning–based approaches to development trajectories of hematopoietic #StemCells.
22
25
135
(1/6) Extremely happy to finally publish our OCAT paper! OCAT is a fast and scalable scRNA-seq integration tool that preserves biological variations while being robust to heterogenous data compared to state-of-the-art methods, such as Seurat, Harmony, and Scanorama.
OCAT: One Cell At a Time, from Wang, Zhang & @BoWang87, a machine learning method for integrating and analyzing single-cell RNA-seq data from multiple sources. It does not require explicit batch correction, and gives cell feature representations.
6
21
131
Excited to see the @nytimes article featuring some recent line of work on foundation models like our scGPT in single-cell analysis! In my chat with @carlzimmer, we dived into how foundation models like scGPT are helpful in advancing the development of “virtual cells.”. So, what’s.
2
18
116
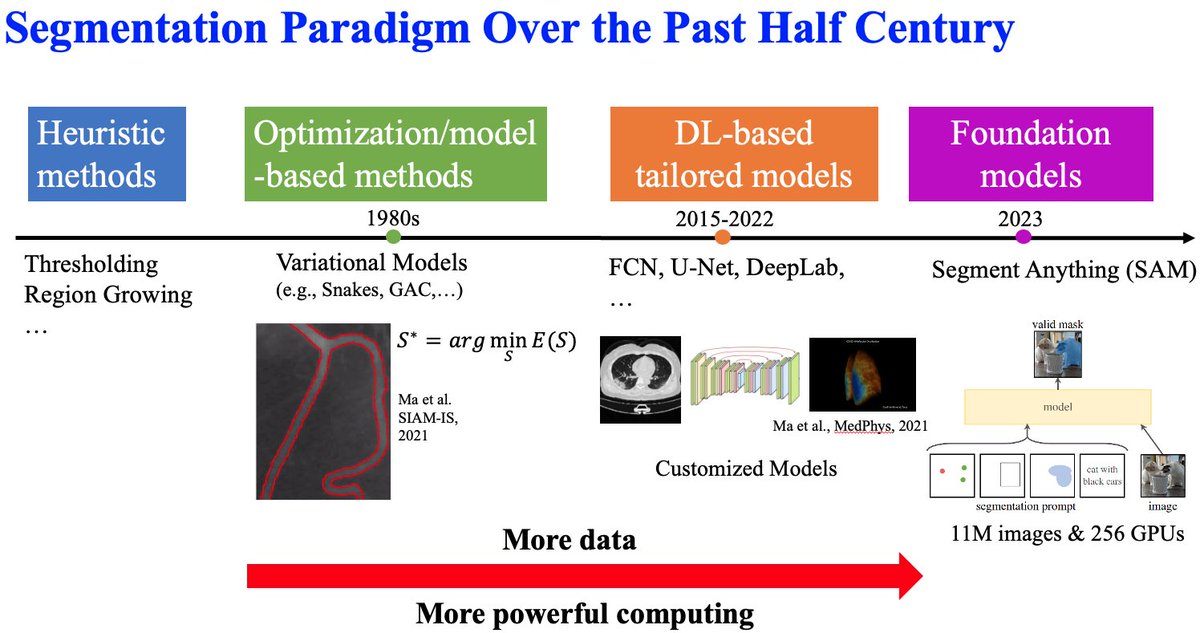
(1/4) Great to see so much interest from the community regarding AI applications for cell segmentation. First of all, Image segmentation has been an active area of research for over half a century, evolving from heuristic methods and model-based approaches to deep learning
(1/5) Attention, Cellular Imaging Enthusiasts! . Thrilled to share that our Analysis "The Multi-modality Cell Segmentation Challenge: Towards Universal Solutions" has been published in Nature Methods (@NatureMethods) 🎉🎉🎉 . Cell segmentation is a critical component of
1
23
112
Revolutionizing modern chemistry and drug discovery, #GNN focuses mostly on single graph representations. Yet, the study of multiple graphs (e.g., drugs) interacting under specific conditions (e.g., cell lines) remains less explored. We present CongFu: Conditional Graph Fusion
3
26
108
When talking about the positive impacts of AI, healthcare and drug discovery always top the list! They're what drives me to get up and make a difference every day. It's an exciting time to be alive!!.
Geoffrey Hinton: 200,000 people a year die of incorrect medical diagnoses in the United States. AI will fix that in the next 10 years.
2
11
105
The partnership between Nvidia and leading biotech and pharmaceutical companies brings together cutting-edge computing with vast biological knowledge and data. Recognizing the unique challenges that differentiate the fields of biology from languages, I am wondering: What else do.
12
9
104
Just to set the record straight, I'm not a UMAP-hater. I believe that both 2D and 3D visualizations are incredibly effective for conveying biological concepts and presenting data to audiences. I acknowledge the limitations inherent in all visualization techniques. Rather than.
Biologist collaborator : What visualization method should I use now? Everyone says UMAP is bad. Me: every visualization method is misleading, but some are useful…. Biologist Collaborator: can you just tell me which one to use? . Me: …. Biologist Collaborator: can you create.
3
14
104
🧬🚀 AI-first biology is hitting its stride, and it’s exhilarating to witness!. A great read from @nathanbenaich at weekend: In just five years, we’ve gone from isolated models to generative AI that’s transforming biology from the ground up. With scGPT,.
2
17
104
Heading to #NeurIPS23 with two papers presented by my students and me:. 1. BLEEP, a pioneering transformer-based model linking H&E images to gene expression in spatial omics. This is a work led by my PhD student, Ronald Xie (@RonaldXie1 ). Paper: .Code:.
2
19
102
Thanks @AIatMeta for properly putting our clinical camel on the map of medical LLMs. We are proud to be one of the earliest open-source medical LLMs, laying the groundwork for many of the subsequent, more advanced LLMs in the field of medicine. More exciting development of
Researchers at @ICepfl & @YaleMed teamed up to build Meditron, an LLM suite for low-resource medical settings. With Llama 3, their new model outperforms most open models in its parameter class on benchmarks like MedQA & MedMCQA. More details ➡️
1
14
103
The structure of the Arc Institute is fundamentally designed for success: significant investment attracts a team of world-class scientists, encompassing both wet-lab and dry-lab experts. By ensuring ample funding and providing robust computing infrastructure, it empowers these.
Arc Institute has been on fire recently. LLM innovation, multiple new gene editing tools, bridgeRNA discovery / elucidation, and more, all in the past few weeks. Really encouraged to see a new academic research funding model proving out, and doing so very soon after inception.
3
7
93
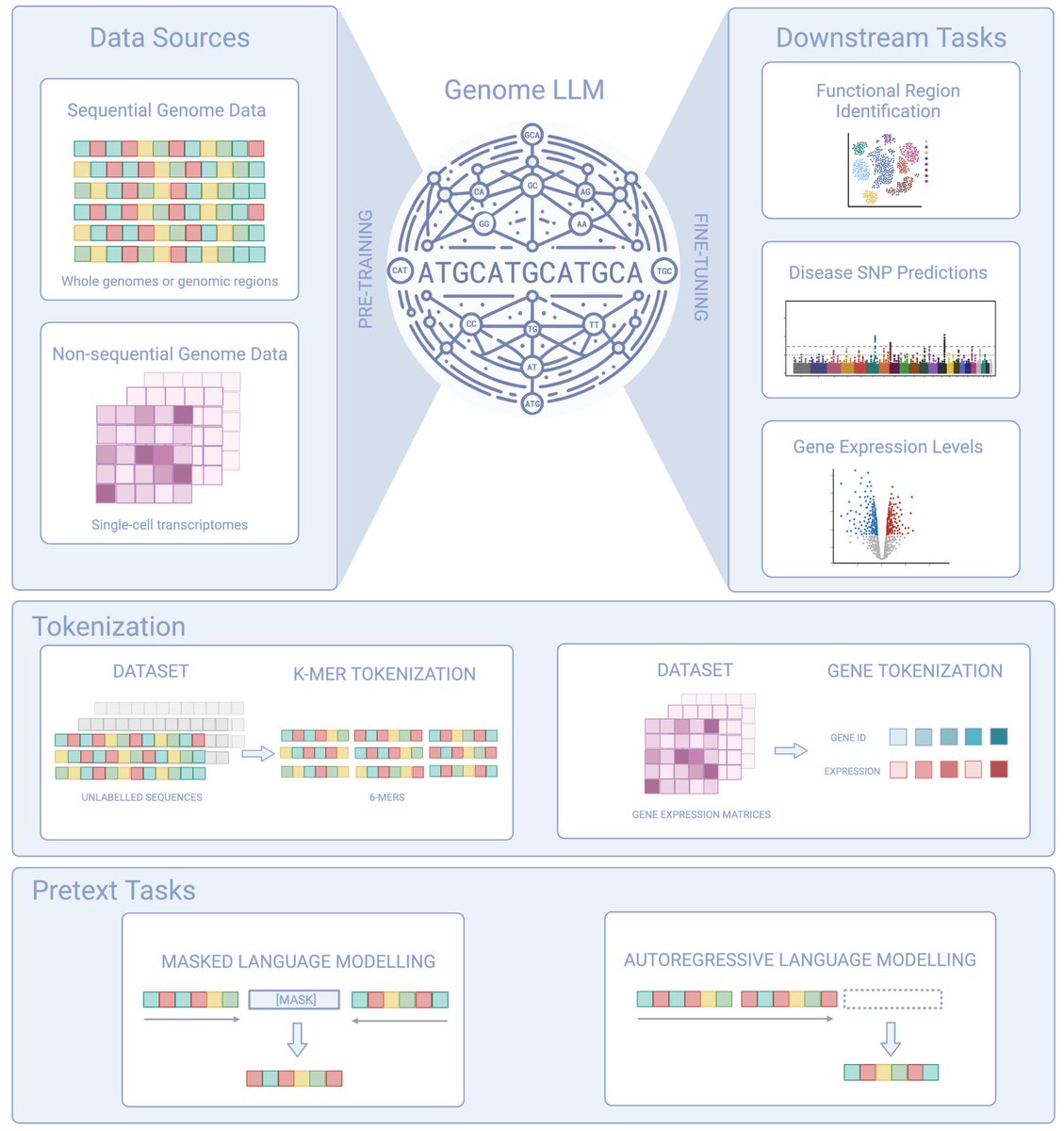
Why RNA Foundation Models?. While protein language models have hit their peak with recent Nobel prize recogitions and DNA language models are getting crowded and messy, RNA models hold unique importance for several reasons:. 1. Predicting mRNA properties is important for.
🚀 We're thrilled to introduce Orthrus 🧬🐕—a groundbreaking mature RNA foundation model designed to push the boundaries of RNA property prediction!. 🔬 What is Orthrus? .Orthrus is a Mamba-based RNA foundation model, pre-trained using a novel self-supervised contrastive learning
0
17
100
Are foundation models all you need for computational pathology?. The recent benchmark paper from Gabriele et al. gave a comprehensive on the existing pathology foundation models on two series of downstream tasks: disease diagnoses and computation biomarkers. here are the.
Excellent work from Gabrielle Campanella, @ThomasFuchsAI @IcahnMountSinai! Benchmarking 8 pathology foundation models including UNI, Virchow and Prov-GigaPath on large scale datasets for clinical diagnosis and biomarker discovery and providing insights into data scaling laws and
1
20
99
Our review paper on transformers in single-cell analysis @naturemethods is out! Great collaboration with @fabian_theis ‘s group! Congratulations to all the co-authors and particularly shoutout to @arturszalata for his leadership in this project! . More details can be seen 👇.
Thrilled to share @fabian_theis lab paper on transformers in single cell omics out in @naturemethods ! Check it out to learn about the latest models, how they are trained, applied, and their limitations. (🧵1/7).
0
18
90
Some personal predictions of the #AIforScience landscape in 2024 🚀:. 1.Expect a rise in non-transformer-based foundation models, like Mamba. 2.Genomic LLMs will gain traction in Pharma for drug discovery and genomic data analysis. 3.Medical LLMs will revolutionize.
1
12
89
It looks like LSTM is making a comeback! This recent paper shows that simply replacing the Mamba module in our U-Mamba with xLSTM can improve medical segmentation performance. Currently, we're working on integrating Mamba-2 into U-Mamba and will update you on which architecture.
Our new xLSTM performs well in vision: and Better than Transformer and Mamba. More applications with xLSTM will come soon.
0
10
88
** 𝙐𝙣𝙫𝙚𝙞𝙡𝙞𝙣𝙜 𝙩𝙝𝙚 𝙋𝙤𝙬𝙚𝙧 𝙤𝙛 #𝘼𝙄 𝙞𝙣 𝙏𝙧𝙖𝙣𝙨𝙥𝙡𝙖𝙣𝙩𝙖𝙩𝙞𝙤𝙣** . Excited to announce our recent publication in #NatureComm titled: “A machine-learning approach to human ex vivo lung perfusion predicts transplantation outcomes and promotes organ
1
18
88
“The next big game-changing revolution is in biology.” . Cannot agree more!. We are at a pivotal moment where vast amounts of biological data are being generated, and large-scale models are increasingly accessible for analyzing omics and sequences. There’s truly no better time.
3
12
88
(1/4)Is image segmentation a solved problem by #AI? maybe for natural images,but hardly true for cell segmentation! To address the gap, we announce the cell seg competition () in multi-modal high-resolution microscopy images in #NeurIPS2022.Join us and RT!.
3
26
81
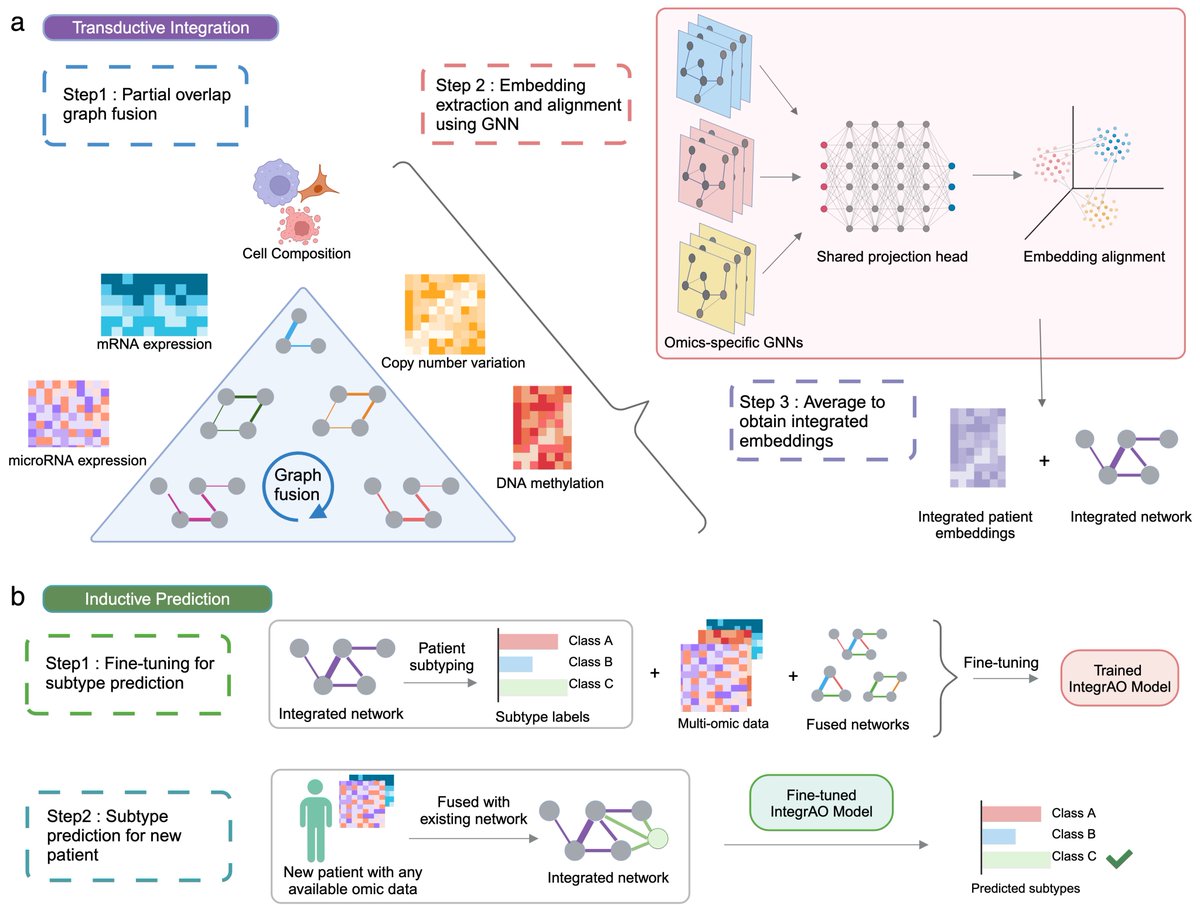
🎉Big cheers for Emily So, an exceptionally talented master student co-supervised by Benjamin (@bhaibeka) and myself, on publishing her first first-authored paper in @NatureMachIntell! TL;DR:. AI models in healthcare are flourishing, but the issue of.
2
20
83
Thrilled to announce that Trevor, a first-year undergrad @UWaterloo and our summer intern at @UHN, clinched 2nd place in the 2023 "A Tumor and Liver Automatic Segmentation (ATLAS)" Challenge! 🎉 (see During his time in our lab, he crafted a DL pipeline
4
4
80
Massive thanks to everyone for supporting our clinical camel! In just a week since going live, we've seen an amazing response worldwide. We're clocking an average of 5000+ daily users on our live demo with over 90% of thumbs-up! - that's incredible! So thrilled to see the.
(Re-)Introducing #ClinicalCamel 🐪: an innovative open-source conversational #LLM engineered specifically for healthcare: . Paper: Model weights in HF: Eval Scripts at Github: Live Demo:
3
18
81
Our fourth speaker, @AI4Pathology from @harvardmed , is talking about recent foundation models in pathology! The workshop is so popular that we even have a line outside of the room .
2
10
79
Wow huge congratulations! AI & biology is really on fire 🔥.
The CZ Biohub Network is pleased to welcome the biomedical & AI expertise of 3 new members to our Board of Directors: .🌟Sam Altman/@sama, @OpenAI CEO.🌟Sam Hawgood, @UCSF Chancellor .🌟Mike Schroepfer/@schrep, Partner @Gigascale. View the full Board: .
2
8
77
Please tune in and it should be fun! Thanks for inviting @Cornell @WeillCornell.
We are thrilled to have Dr. Bo Wang (@BoWang87) talking about biomedical foundation models in our February seminar. Register here to hear Dr. Wang’s amazing work.
3
17
74
#DiffusionModels have revolutionized generative AI, particularly in nature imagery. But how about medical images?. Our recent #MICCAI2023 paper, "Pre-trained Diffusion Models for Plug-and-Play Medical Image Enhancement", addresses it. Medical image enhancements often rely on.
0
15
76
Very thrilled to witness Toronto General Hospital at @UHN secure a top 3 global ranking! This outstanding accomplishment fills us with pride, especially knowing our AI innovation in lung transplantation played a pivotal role in achieving this success!Congratulations!.
2
8
73
Our perspective of how to build the #AI virtual cell is out in @CellCellPress ! This is an amazing vision led by @cziscience ! . Now it is time to get the work started! #scGPT 🔥🔥🔥.
Towards the #AI virtual cell, which would represent one the most extraordinary achievements in life science.@CellCellPress @StephenQuake @czi @cziscience @_bunnech
1
15
74
Honored to give the keynote for the Bigelow Lecture Series by @UofT Surgery @uoftmedicine! Discussed how generative AI can transform healthcare with our lab’s projects: Clinical Camel, MedSAM, and scGPT. Grateful for the amazing turnout from surgeons and trainees. Inspired by
0
10
70
This seems out of control 🙈🙈
What is the likelihood of this happening: . P(doom) = P( author i did not read the paper)^ 5 * P( reviewer j did not read the paper) ^ (2 or 3) * P(editor did not read the paper) * P( journal proofreading staff did not read the paper) . And yet it still happened 😂😅
9
9
72
Grateful to @globeandmail for featuring our progress in #AI for #singlecell analysis. A round of applause for @garybader1's outstanding leadership in orchestrating and guiding numerous extensive cell atlas projects! @UofT_TCAIREM @UofTCompSci @UofT_LMP @DonnellyCentre.
Scientists and AI are on a quest to map every cell in the human body, charting where and why disease happens
1
15
65
Late to the discussion, but I always value insights from @mo_lotfollahi! 🚀 Great seeing more evaluation papers on foundation models. Check out this recent one from other group too: While any comments I make about scGPT should be taken with a pinch of.
(1) This is an amazing paper on the limitations of (current) single-cell foundation models "Our results indicate that both Geneformer and scGPT exhibit limited reliability in zero-shot settings and often underperform compared to simpler methods.".
3
10
70
Thanks for the shoutout @UofTCompSci ! Congratulations to all the award recipients!.
U of T AI researchers Amir-massoud Farahmand, Gennady Pekhimenko and Bo Wang are recent recipients of Ontario Early Researcher Awards for projects on reinforcement learning; computer systems; and deep learning tools for heart failure prediction.
8
5
72