Hani Goodarzi
@genophoria
Followers
5K
Following
9K
Media
129
Statuses
4K
Core Investigator @arcinstitute | Associate Professor @Ucsf_Biochem @UCSFurology @UCSFCancer | {Computational, Cancer, RNA} biologist
San Francisco
Joined June 2009
I am deeply grateful to the Vilcek Fdn @Vilcek for this recognition. Immigrants are and have long been the backbone of science and technology in the US. I am surrounded by visionary immigrants, and I am excited to be their representative for today.
🔬🧬🧪Congratulations to the recipients of the 2022 Vilcek Foundation Prizes in #BiomedicalScience. Vishva M. Dixit - @dixitvishva - receives the Vilcek Prize. @Landry_Lab, @genophoria, and @harriswangnyc receive Vilcek Prizes for Creative Promise.
63
21
366
A month ago we @vevo_ai announced that we have generated the largest single-cell perturbation atlas in history, Tahoe-100M. Today, we announce that we will fully open-source Tahoe-100M in Feb, as part of a collaboration with @NVIDIAHealth to train cell state models.
3
55
378
I am excited to join all the amazing investigators and scientists at @arcinstitute! Can't wait to do some cool computational and technology-centric team science! And looking forward to joining forces with the innovation and ignite awardees to expand our community!.
We’re thrilled to welcome Hani Goodarzi @genophoria, Associate Professor of Biochemistry & Biophysics @UCSF, as our newest Core Investigator. His groundbreaking research uses machine learning models and experimental frameworks to dig into cancer progression and metastasis.
53
15
295
Super excited to see the launch of @exaibio. When @lisacfishsci and I started our search for orphan RNA species (oncRNAs), never did we imagine it will lead us here. Now some of the smartest people I know will be working on this problem! @babak_alipanahi @lisacfishsci.
The Vilcek Foundation congratulates Hani Goodarzi and the team @exaibio on securing $67.5M to finance a new RNA-based platform for early #cancer detection. @genophoria is the recipient of a 2022 #VilcekPrize for Creative Promise in Biomedical Science.
43
18
280
Excited to see this out after such a long time. Debt of gratitude to all the colleagues and collaborators who helped with this over the years: @SohailTavazoie and the lab @RockefellerUniv @hsnajafabadi Claudio @FKHM.
48
61
281
Excited to announce Vevo’s launch! What started as coffee chats with @kevansf close to 7 years ago has now given us an opportunity to rethink how we do drug discovery.
Vevo Therapeutics has officially launched out of stealth today with an oversubscribed and upsized $12M seed financing. We are so excited to reveal what we have been building behind the scenes! 🧵1/6.
42
22
221
@iamjohnnyyu, @nalidoust, @kevansf, and I are excited to announce a historic achievement by our team at @vevo_ai: Tahoe-100M, a 100M single-cell transcriptomic atlas, measuring how thousands of drugs impact cells from 50 diverse cancer lines.
1
41
179
I am late to my own party, but I wanted to write a short 🧵 to put this talk in context. First of all, the @TEDTalks team was amazing; there is so much that goes into these seemingly short talks. It was an amazing experience and I made some new friends.
4
20
143
Thanks to @AACR and @MPMCapital for the support. I am extremely grateful to @AACR for their crucial and continued support over the years.
Kudos to Hani Goodarzi @genophoria recipient of AACR-MPM Oncology Charitable Foundation Transformative Cancer Research Grant, and thanks to @AACR @UBS @MPMCapital for funding high-risk/high-reward #CancerResearch.
21
10
141
A long time ago, @ElementoLab and I came up with our solution to the gene-set enrichment analysis. We used mutual information to measure the association between gene-set membership and input. After a decade, a talented undergrad spearheaded an update:
4
24
129
The first preprint of 2025! Together with @khorms, @halfacrocodile, & our amazing team, we are excited to share PARADE: an AI framework for designing mRNA UTRs with enhanced cell-type specificity & stability.
4
29
128
Check out our latest paper in @NatureCellBio exploring the link between alternative polyadenylation, translation efficiency, and metastasis.
Links between translation and alternative polyadenylation in metastasis? The story is out today! A great collab between @genophoria, @AndreiGoga_SF and @FKHM labs! Shout out to co-authors @Gentremet, @JulianeScience, @lisacfishsci among others!
7
21
122
Check out our latest research by @MNaghipourfar and others in our team on building language models of coding sequence. In this preprint, we introduce cdsFM, a suite of codon-resolution language models!👇.
3
24
120
Took us a minute, but I am happy to share the final version of this study, now published at @NatureCancer, on behalf of Bruce and Kristle. We have it all, regulatory RNA interactions, oxidative stress/ferroptosis, and metastasis:
Excited to share our second preprint of the week, courtesy of our former lab specialist Bruce Culbertson and our resident specialist Kristle Garcia, with help and support from many others, most notably Josh Rabinowitz’s group!
11
15
113
Our first paper and really great work by Dr. Lisa Fish (a postdoc in my lab) and @at__steven (former HHMI med student fellow) and great collaboration with @AndreiGoga_SF. Below is a thread summarizing our work if you're interested in learning more.
3
28
104
Our latest from the indefatigable @iamjohnnyyu in collaboration with @JswLab and @kevansf. Meet GENEVA, which enables simultaneous phenotyping and profiling of cancer cell drug responses at scale; both in vitro and in vivo across a variety of models:
2
23
114
Genomic interval tools, like bedtools and bedops, have formed the backbone of our analytical pipelines for many years now. @noamteyssier has now developed a new package called 'gia' that is order of magnitude faster! Check out our preprint and take gia for a spin.
1
25
104
I have been thinking about the best approaches for building predictive models in liquid biopsies for many years now! Orion, which was built by the @exaibio team with input from our advisors @ElementoLab and @james_y_zou is the culmination of that effort.
2
30
97
A great start to 2024! Thank you for the shoutout @EricTopol. You said it all!.
How do you go from 9 million molecules to 8 which appear effective against a micro RNA driver of breast cancer?.Like this.@Patterns_CP #AI @genophoria @UCSF @UCSFCancer
8
10
91
Really excited to see this out. Congratulations to Matvei and everyone who helped us along the way! Shout out to @LukeGilbertSF and @yifan_ucsf . without their help, this wouldn't have been possible.
5
11
92
The Computational Tech Center at @arcinstitute is recruiting an experienced ML Engineer to work on implementing and deploying frontier models for biology. Come join @yusufroohani, @BrianHie, @a_dobin, @pdhsu, and I as we leverage AI to model biology, from genomes to cells.
2
24
86
The final version of is now published: Congratulations @khorms57 and our team across 3 groups, ours, @FKHM, and @halfacrocodile!.
Thanks to @vishvak2000 we are also releasing an RNA binding protein (RBP) browser for dynamic visualization of RBP-RBP interactions in our preprint (1/7)
1
23
85
Y'all, check out our latest preprint on determinants of small RNA release. We used machine learning to show much of the information is encoded in the primary RNA sequence and can be leveraged to predict which small RNAs are effectively secreted. See this thread from @BaharZirak.
Super excited to share the results of our highly collaborative project co-led by @MNaghipourfar, Ali Saberi, @DelaramPB, Amirhossein Zarezadeh and supervised by my incredible mentor @genophoria.
2
12
80
I am reviving this thread to announce the final publication of this preprint. Almost a full year after the preprint was first submitted! A clear example of how biorxiv expedites dissemination of science.
Our first preprint, in which we describe a regulatory function for nuclear TARBP2. Comments & suggestions are welcome.
6
13
73
I came to this country 15 years ago, and it is unbelievable to me that we haven't managed to improve this system even an iota in these years. It is not just a cruel system, it is also self-defeating. Please read this thread and tag your senators. @AlexPadilla4CA @SenFeinstein.
I’m quite used to the cruelty students can face when they apply for a US visa but this one broke me. We offered admission to a stellar, talented & hardworking student. After months of work and hundreds of dollars, an embassy officer saw him for 5 mins & said no. why? ….
0
11
70
I'm super happy that the UCSF magazine has covered this story. @FaranakFattahi 's work is nothing short of inspirational. Cracking the Coronavirus Gender Mystery
2
16
69
The first time I really thought about RNA was when I read the major transitions in evolution, and I have been hooked ever since. Happy #RNAday!.
1
0
68
Thank you for having us @arcinstitute!.
☀️It’s a great week at Arc as we welcome our new Core Investigator @genophoria and his lab, and our first Science Fellow @zhou_jingtian! We’re proud to add these talented minds to our ranks, bringing with them a multidisciplinary approach to human disease research. Welcome, all!
1
3
68
3300 subjects across 8 cancers; overall stage I sensitivity of 87% at 95% specificity in the held-out test set. 88% tissue-of-origin accuracy! Watching the @exaibio team do their magic has truly been a privilege.
Exai announced compelling new data at #ASCO23, demonstrating strong performance in stage I detection of multiple cancers including breast and prostate cancers, overcoming early-stage detection limitations of other blood-based approaches.
3
4
64
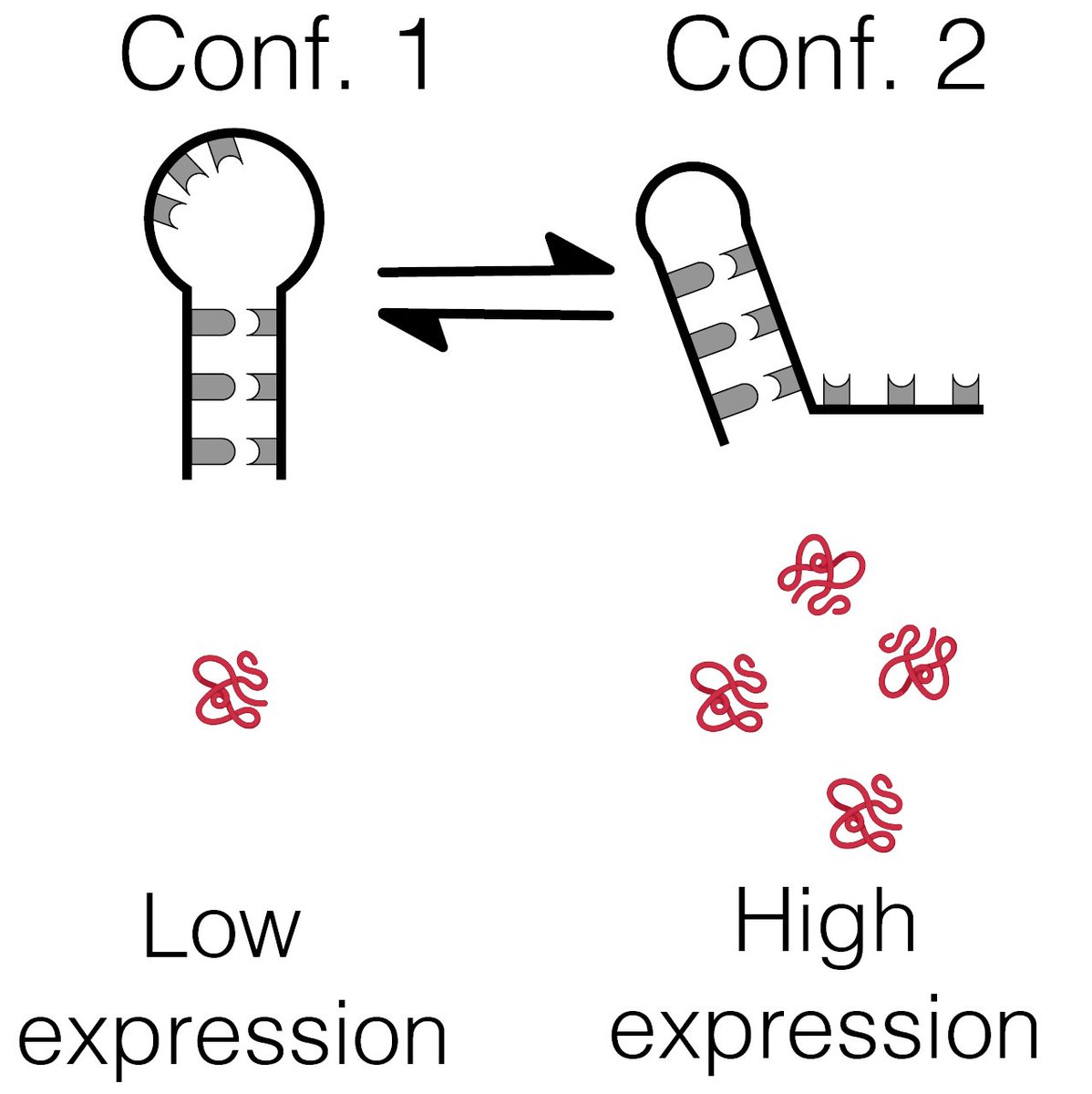
Another movement from @khorms57's magnum opus in our lab. A rigorous attempt at systematically annotating functional RNA structural switches that show conformation-dependent regulatory functions 👇.
RNA folding affects gene expression, but this link is not well explored in Eukaryotes. We researched RNA structural switches and discovered hundreds of them in the human transcriptome. We even visualized some of their 3D folds with CryoEM. Thread below:
3
7
65
Hey y’all, check out our latest preprint and this fantastic Tweetorial from our senior postdoc @a_navickas! h/t @a_navickas @Gentremet @lisacfishsci @AndreiGoga_SF @JulianeScience @FKHM.
Excited to share the latest results from the @genophoria lab & a great collaboration with @AndreiGoga_SF and @FKHM labs! Shout out to the amazing co-authors @Gentremet, @JulianeScience, @lisacfishsci and many more!
1
10
61
Check out this beautiful in vivo perturb-seq study from @LukeGilbertSF, @LabRaleigh, and Bill Weiss at @arcinstitute and @UCSFCancer!.
In vivo perturb-seq of cancer and immune cells dissects oncologic drivers and therapy response #bioRxiv.
0
8
57
@CarolynBertozzi talking about bioorthogonal chemistry at @arcinstitute Investigator retreat. #arclife
0
0
58
We started this #RNAyear by saying goodbye to two longtime amazing postdocs: @a_navickas who is off to Institut Curie to start his own "RNA in cancer" lab! And @MKarimzade who is joining @exaibio to build ML models for disease detection. Certainly a bittersweet moment for our lab.
2
1
57
Super exciting news! I am happy that our oncRNAs are living their best lives at @exaibio! I am also excited for this strong commitment to breast cancer. Breast tumors don't shed a lot of DNA and DNA-based tests show poor performance at low burdens. OncRNAs will change that. .
Based on initial positive data, Exai is proud to announce expanded participation in the I-SPY 2 Trial to advance our novel RNA-based liquid biopsy platform for breast cancer. s-Novel-RNA-based-Liquid-Biopsy-Platform-in-Breast-Cancer.
1
0
54
We attempted the whole online class thing for our systems biology course. Turned out better than expected! #scRNAseq @mattjonesucsf
1
6
53
Please take a few minutes out of your busy day and read our preprint with the @FaranakFattahi lab. Hi-quality in vitro drug screen, supplemented by in silico GCNN screens, followed by targeted validation and clinical confirmation.
Our preprint on COVID-19:.•Antiandrogenic drugs reduce ACE2.•higher free androgen predicts poor outcomes in men.Is this why men are more affected & kids are protected?.Big thanks to @genophoria @UCSF, @xaniarg @YaleIMed, @pnatarajanmd @MassGeneralNews.
1
8
53
Shout out to @sinabooeshaghi for showing up to our systems biology class (via zoom) and giving one of the best presentations of the whole course! Also I’m told he has some exciting stuff brewing. so, follow him and stay tuned!.
1
1
52
I am devastated by this news. She was a fantastic mentor. so kind and generous with her time and resources. A great loss for us at UCSF and the entire cancer research community. She will be missed.
Zena Werb. Today we have lost one of UCSF’s most beloved faculty. Zena Werb died suddenly this morning. A member and VC of the Department of Anatomy, for almost 40 years, Zena was an outstanding scientist. She was one of the world’s leading experts on ECM on cancer development
0
7
48
Estimate RNA stability changes from RNA-seq alone. Inspired work by @hsnajafabadi ! Excited to have been part of it.
1
22
45
For all the RNA peeps out there attending #RNA2020 online. If you are interested in the role of regulatory RNA structural elements in RNA splicing and disease, please check out my talk (15). Happy to answer questions here or any other platform.
0
4
46
Please check out our latest preprint, together with @FKHM, and @halfacrocodile, along with a detailed Tweetorial from Matvei. Learn about post-transcriptional regulatory modules and how we went about identifying them:
Can’t wait to share the splendid results of the collaboration between the incredible labs of @genophoria, @FKHM, and @halfacrocodile! Tl;dr: we've explored the code on how RNA-binding proteins team up to regulate their target RNAs. Read on for more! (1/15)
2
8
46
@dsboothacosta Funding for grad students (stipend+tuition+research) should be separated from our grants and be b/w the program & students. Then students can join the labs that best suit them, vs just well-funded labs. It also clarifies our roles as mentors/advisers & not bosses.
1
2
43
It was a lot of fun talking to Frontier Science about my thoughts on multidisciplinary research at the intersection of computational and experimental biology, our work in this space, and our path towards translation.
Frontier Science 🧫 - Modeling Oncological Systems 🧬 w/ Hani Goodarzi - Associate Professor @UCSF. Learn more about Goodarzi's (@genophoria) work & the below.
0
1
49
Check out our latest preprint with the @hsnajafabadi lab! Leveraging matched short- and long-read RNA sequencing to achieve optimal isoform quantification.
What is the best strategy to quantify mRNA isoforms? Short-read RNA-seq to achieve high depth? Or long reads to get unambiguous read-isoform assignments? In our latest work with @genophoria, we show that what we really need is to combine both! (1/n)
1
6
48
Late better than never! We celebrated PAW with our amazing postdocs working on site. With @a_navickas & Heather who basically run the lab, and welcoming Benedict and Siyu who just joined recently. Looking forward to next year and all the awesome science they are doing!
1
0
47
Celebrating another year with our awesome postdocs. My life would be insanely more difficult without them! From right: Drs. Fish, Navickas, Asgharian, Baygi. #ucsfNPAW #ucsfpostdocs #NPAW #NPAW2019
0
0
45
Had loads of fun with @AndreiGoga_SF, @girlsinscience_ , and the UCSF BOP family at the Tri-Valley SOCKs breast cancer walk yesterday! Sad we said goodbye to this event! #breastcancer #SteppingOutforCancerKures
2
0
44
Early access to a perspective by Peter Walter and @MullinsLab "From Symbiont to Parasite: Evolution of For-Profit Science Publishing". p7: "what remains puzzling is the lack of more widespread anger in our communities regarding the degree of exploitation".
0
18
37
pyPAGE is now officially out! New spin on old problem.
A long time ago, @ElementoLab and I came up with our solution to the gene-set enrichment analysis. We used mutual information to measure the association between gene-set membership and input. After a decade, a talented undergrad spearheaded an update:
1
6
40
@s_tavazoie @PNASNews I found this sketch from 10 years ago when @s_tavazoie first told me about (a version of) this idea! Few in this day and age show this level of stick-to-itiveness! Congratulations Panos and the @s_tavazoie lab.
0
1
36
Had a blast in Bergen! Come for the views, stay for the science. Thank you @Halberglab1 for the invite. Really, A gem of a place! Extra points for the pride celebration!
1
0
41
Check out this tour de force from the Fattahi lab! It has it all… platform building, basic biology, drug screening, functional validations….
We made enteric ganglioids from hPSCs, showed they’re similar to human ENS, used them to find neuromodulators that regulate GI motility and demonstrated their engraftment potential in mouse colon. Check out our new preprint by @homamajd et. al.
0
4
42
As our NextGen grant for transformative cancer research comes to an end, I had an interview with @AACR on the impact of this grant on our research. Spoiler alert: we may have not transformed cancer res, but certainly tried. and the night is still young.
0
2
41
It was so much fun having @gdb at @arcinstitute! Onwards and upwards!.
During my sabbatical, one project I really enjoyed was training DNA foundation models with the team at @arcinstitute. I've long believed that deep learning should unlock unprecedented improvements in medicine and healthcare — not just for humans but for animals too. I think this
1
1
42