Soufiane Mourragui
@souf_mourra
Followers
499
Following
8K
Media
2
Statuses
587
ML @ Ensocell | Previously @_Hubrecht @AlexandervanOu1 @TUDelft @NKI_nl @Mines_ParisTech @psl_univ | Comp-Bio, Single Cell, Oncology
Cambridge, United Kingdom
Joined October 2017
I am thrilled to share our latest pre-print comparing cell lines and tumors at single cell resolution using alignments of deep generative models. @wesselslab_nki
@oncodeinstitute @EEMCS_TUD
biorxiv.org
Preclinical models are essential to cancer research, however, key biological differences with patient tumors result in reduced translatability to the clinic and high attrition rates in drug develop...
1
9
33
🎉Check out our paper on computational #antibody design guided on properties. Now published at @PRX_Life!👇
Researchers improved a deep generative diffusion model to guide #antibody design. Their approach identified complementarity-determining regions in antibodies with properties that would be suitable for clinical applications. #MachineLearning #MLSB 🔗 https://t.co/mhXcLfBowk
0
2
5
Super happy we managed to put this together. Such an awesome group of speakers and a lovely location, you don't want to miss it!
Come join us for a 1w @ISBSIB spatial omics workshop in beautiful Lausanne 🇨🇭 > https://t.co/O9EAifjBne < featuring @LarsBorm @ahmedElkoussy @markrobinsonca @YvanSaeys @RaSaghaleyn @AnnaCSchaar @MarcoVarrone & more - deadline: Nov 1st.
0
2
10
We are excited to share our new research article, in which we, for the first time, identified a human intestinal reserve stem cell, namely tuft cells, by combining novel organoid models and single-cell genomics (link: https://t.co/ZN5mqLMFWL).
10
9
73
Please have a look at our g2g framework on aligning cell trajectories developed by Dinithi Sumanaweera, Chenqu Suo @ana_cujba and others in the team! Congrats! @sangerinstitute @SCICambridge @Cambridge_Uni
Genes2Genes is a Bayesian framework developed by @Teichlab for aligning single cell pseudotime trajectories at the per gene level. https://t.co/dfvQBm14p4
1
10
63
Ready to establish your own research group at the Hubrecht Institute for molecular and developmental biology? We’re seeking a tenure-track group leader to develop an innovative research line within our vibrant scientific community. Learn more & apply! https://t.co/hXb5jz43oG
1
45
63
I’m thrilled to announce that I will join the Institute of Artificial Intelligence at the Medical University of Vienna (@MedUni_Wien), led by @BockLab, as a tenure-track assistant professor starting December 2024! Check out our website and open positions
ai4biomedicine.org
Academic lab for mechanistic machine learning and AI models of cells. We focus on cell biology captured by single-cell and spatial omics.
32
28
267
Can cells adapt to aneuploidy? Our new study reveals that reducing chromosomal instability and inflammation is key to adaptation. Thanks for everyone that contributed! @_ms03 @marsotordlt @JonneRaaij @MedemaRene
https://t.co/ErzsAhgYqj
embopress.org
imageimageRPE-1 cells adapt to aneuploidy in vitro, a process characterized by reduced CIN and inflammation. This study shows that this can be achieved through metabolic rewiring and KRAS amplifica...
0
8
31
Don't miss poster P296 today at @ECCBinfo if you want ro know more about unpaired multi-modal data integration from @JulesSamaran !!!!
After a very constructive back and forth with editors and reviewers of @NatureComms, scConfluence has now been published @LauCan88 @gabrielpeyre ! I'll present it this afternoon at the poster session of @ECCBinfo (P296) Published version:
0
2
9
Excited to share this new pre-print!
gsQTL: Associating genetic risk variants with gene sets by exploiting their shared variability https://t.co/a8SzCUsPWC
#biorxiv_bioinfo
0
3
5
Extremely honored to be invited to write about scEdU-seq as Tools of the Trade article in @NatRevMCB . Many thanks to @lisa_heinke for the invitation and opportunity! 🎉 https://t.co/o8jfVlTCiP
0
6
24
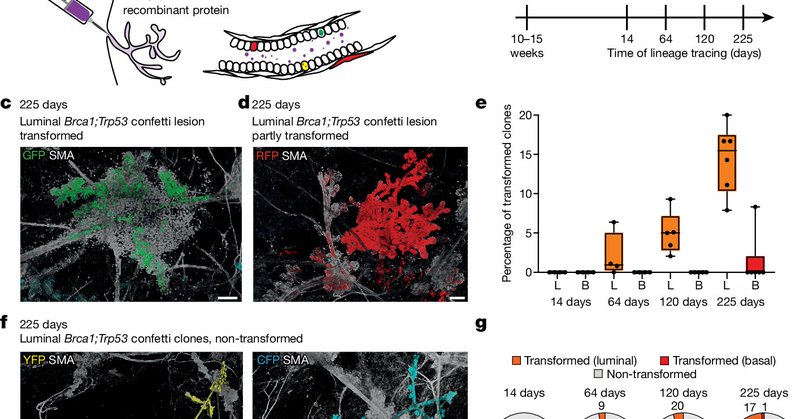
Super happy to share that our paper is published in @Nature today! We've discovered how the breast is protected against tumor initiation—though it comes at a cost. 📰Read all about it here: https://t.co/VmJdPNt7AW 🏥@vib_ccb @Onco_KULeuven @NKI_nl @vanRheenen_Lab @BenSimons_Lab
nature.com
Nature - The authors use lineage tracing to map the fate of wild-type and Brca1−/−;Trp53−/− cells in the adult mouse mammary gland, identifying three layers of protection...
21
50
251
Have you ever wondered about how the genomic position of regulatory elements impact their functional output? Check out our latest pre-print from the @bvanssteensellab.
biorxiv.org
Genes are often activated by enhancers located at large genomic distances. The importance of this positioning is poorly understood. By relocating promoter-reporter constructs into >1,000 alternative...
2
21
61
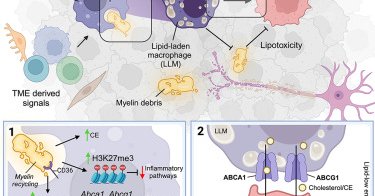
Paper 🚨! Ecstatic, proud, grateful …Years of dedication from the whole lab, with the exceptional @djkloosterman and the one of a kind #JohannaErbani at its helm. What a journey all the way to @CellCellPress! How did it start (& end)? A 🧵1/n https://t.co/d1SPgjFRDr
cell.com
Recycling of cholesterol-rich myelin debris by macrophages leads to the emergence of lipid-laden macrophages (LLMs) in glioblastoma. LLMs support glioblastoma malignancy through transfer of myelin-...
51
92
399
One of my mentees just completed his second project with me: SciKit-Jax: https://t.co/nDPHmfEQCR Scikit-Learn does not leverage GPUs by itself and offers less flexibility in the model design. Scikit-Jax solves these! He will polish this to accommodate multi-GPU setups once we
github.com
Your favourite classical machine learning algos on the GPU/TPU - LiibanMo/scikit-jax
8
19
138
"Transformers are Universal In-context Learners": in this paper, we show that deep transformers with a fixed embedding dimension are universal approximators for an arbitrarily large number of tokens. https://t.co/gEPoC3W2gC
18
319
2K
Due to their unique exploration challenges, the data from emerging #singlecell #epigenomics protocols can be difficult to work with. We present sincei, a toolkit for exploring sc(epi)genomics data directly from BAM files, using bash one-liners or #python
biorxiv.org
Emerging single-cell sequencing protocols allow researchers to study multiple layers of epigenetic regulation while resolving tissue heterogeneity. However, despite the rising popularity of such...
1
6
14
🎉 New preprint! https://t.co/XXspy6nlwX STORIES learns a differentiation potential from spatial transcriptomics profiled at several time points using Fused Gromov-Wasserstein, an extension of Optimal Transport. @gabrielpeyre @LauCan88
2
22
70
Our latest: The innate immune landscape of dMMR/MSI cancers predicts outcome of nivolumab treatment: results from the Drug Rediscovery Protocol 4 key novelties below 👇
1
4
8
Happy to announce that in September 2025, I will open my research laboratory at @NYU_Courant 🏢🔬. As an assistant professor of CS & Biology 💻🧬, I will carry on my work on advancing ML research for molecular biology, and apply those methods for scientific discoveries 🌟🔍.
56
31
479