lewis martin
@lewischewis
Followers
1K
Following
13K
Statuses
602
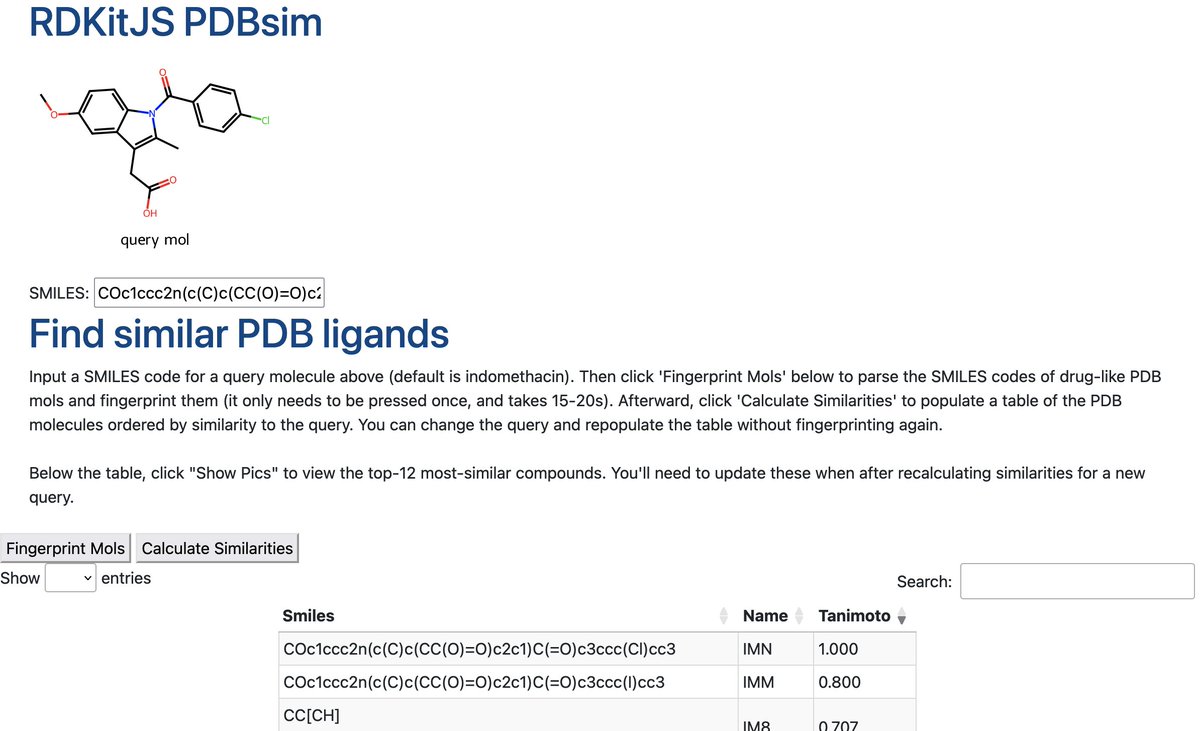
RT @RowanSci: RDKit is the standard software for cheminformatics, but RDKit doesn’t integrate naturally with high-accuracy chemistry simula…
0
6
0
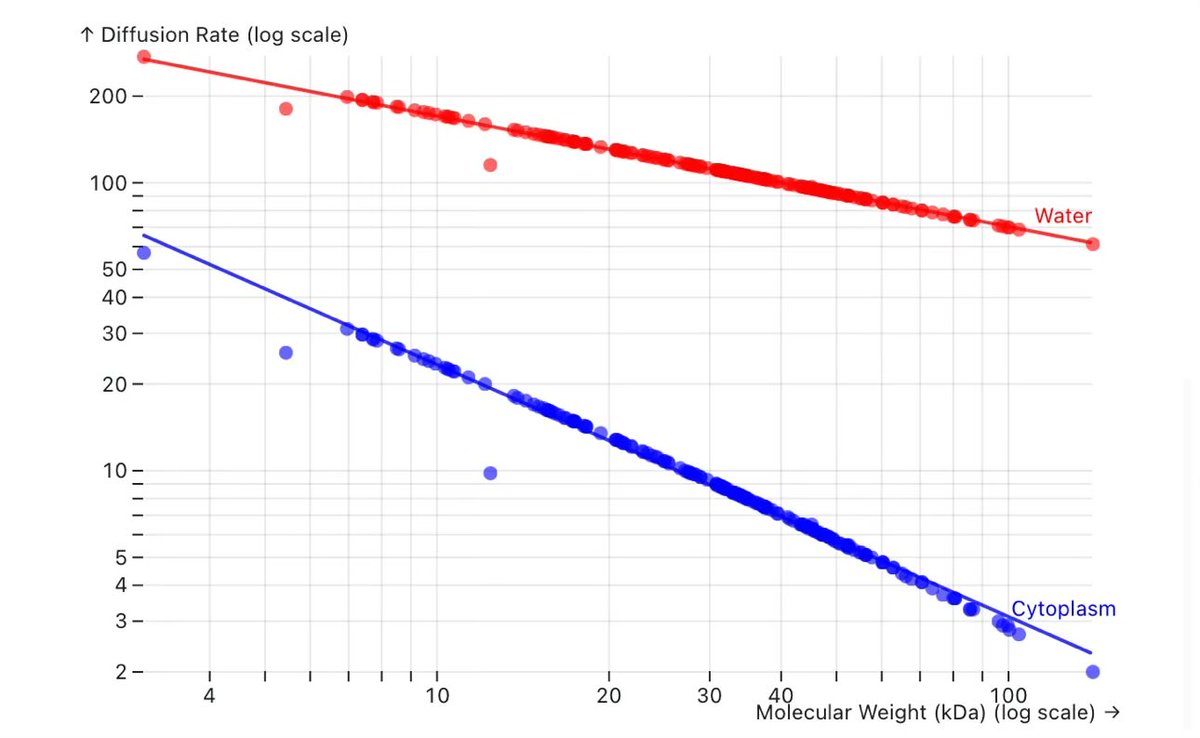
We all know about cell potency dropoff due to permeability, efflux, or plasma binding. This adds another angle: diffusion inside the cytoplasm is 10X slower than water, so your effective Kd in the cell is worse! Off-rate remains the same, but fast on-rate requires fast diffusion
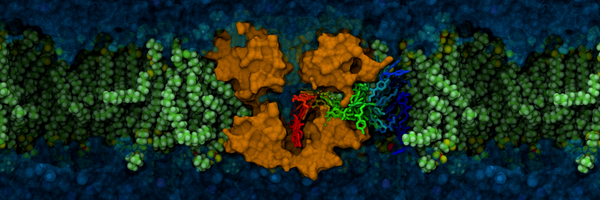
New Blog: A protein takes 0.01 s to traverse E. coli's diameter. The same protein takes 4 min to move 1 mm and >6 hours to move 1 cm. The farther a molecule must move, the less likely it will get there. This is partly why cells are very tiny — it makes collisions more likely.
0
0
14
RT @ProfvLilienfeld: Unlock the mystery of entropy with my latest #StatisticalMechanics lecture now on YouTube! 🌪️📷 We delve into how entro…
0
3
0
RT @CorinWagen: New preprint! With @JosephJGair1 + the Gair lab @MSUChem, we built a set of strained conformers for benchmarking DFT functi…
0
24
0
@exolabs @GoogleDeepMind @exolabs are these connected to the same wifi network, or do you somehow create a direct wired network between them? Does using wifi introduce much latency? TY!
1
0
0
@TimothyDuignan question re: the K+ channel example. How does one verify the NNP sim is a correct novel prediction, or a quirk outside of the applicablty domain? Additive MD has approximations that lead to spurious results too, but theyre physics based and they’re often explainable and fixable
3
0
4
RT @charles_irl: Now upgraded to a fully-fledged @modal_labs example! Fold proteins at scale, without getting a second PhD in Kubernetes.…
0
2
0
RT @BiologyAIDaily: Predicting multiple conformations of ligand binding sites in proteins suggests that AlphaFold2 may remember too much @P…
0
17
0