Sho Suzuki (he/him)
@ShoSuzuki20
Followers
561
Following
3K
Media
53
Statuses
446
Assistant Professor at School of Biological Sciences, Nanyang Technological University, Singapore. NRF fellow, Postdoc@Cornell, PhD@Tokyo Tech
Joined December 2019
I'm absolutely blown away by this incredible research! SNARE insertion into an autophagosome, temporally regulated by PI4P, can be reconstituted in vitro. Huge congratulations to @SaoShinoda, our Weill Institute star @MasaakiU5, and all the team! 🌟.
Syntaxin 17 recruitment to mature autophagosomes is temporally regulated by PI4P accumulation #biorxiv_cellbio.
1
5
21
I am excited to share my postdoc work as a preprint. Mvp1/Snx8 mediates retromer-independent recycling.
A PX-BAR protein Mvp1/SNX8 and a dynamin-like GTPase Vps1 drive endosomal recycling #biorxiv_cellbio.
2
4
18
Fantastic work from @MizLabUT!.
0
1
10
This is a really cool paper. New lipid transfer proteins for endosome-to-Golgi traffic.
SHIP164 is a Chorein Motif Containing Lipid Transport Protein that Controls Membrane Dynamics and Traffic at the Endosome-Golgi . #biorxiv_cellbio.
0
2
9
Good words, Jeff!.
Wise words from Emr &co (@Cornell):."contact sites are useless without proteins that facilitate some sort of transfer, sensing, or communication between the membranes. Some tethering function is likely inherent in the biology of many of these facilitators".
0
1
6
Great work, Lu! Congratulations!.
Zhu et al @CornellMBG show that Rcr1 & Rcr2, two adaptors for the E3 ligase Rsp5, are sorted to distinct locations. Exomer sorts Rcr1 to the plasma membrane and the upregulation of Rcr1 via the Calcineurin signaling pathway maintains cell integrity
0
0
6
Atg9 is a scramblase! Beautiful studies revealed the most surprising finding in the autophagy field. Congrats Matoba-san and Noda-san!.
"Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion" by Kazuaki Matoba, Nobuo N Noda et al. , Nature Structural & Molecular Biology 26 Oct. 2020.
0
0
5
Congrats! @MasaakiU5!.
Congratulations to our newest postdoc, @MasaakiU5, for being awarded an HFSP Postdoctoral Fellowship! @weillinstitute @CornellChem
1
0
5
Fantastic review! Absolutely recommend it! 🙌.
How do #organelles acquire #membrane #lipids? Our review in @AnnualReviews of Cell and Developmental Biology defines a family of bridge-like lipid transport proteins that control membrane area and so much more! Check out the ahead of print version now. @YaleCellBio @AGuillenSaman.
1
0
5
Takeshi Noda, who was a postdoc in Emr lab and a PhD student in Ohsumi lab, discovered yeast mTOR downregulates autophagy. This is a really good news article.
Yoshinori Ohsumi, Takeshi Noda and colleagues at the National Institute for Basic Biology in Japan discovered that the Tor protein controls the breakdown process of autophagy in yeast cells. @Spieringmj writes about their classic paper in @jbiolchem:
0
1
3
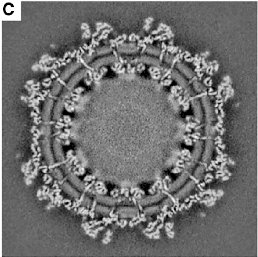
This is amazing.
This is #ChikungunyaVirus imaged by cryo-electron microscopy 🔬The level of detail we can now get blows me away 😱 Want to know why I love science? LOOK HOW AMAZING IT IS. You can very clearly see the E2 protein crossing the lipid bilayer 👨🏼🔬. Ref on sub tweet. #virology #scicomm
0
0
3
@My_Liberty_ コーネル大学で研究員を6年やっているものです。この度は誠におめでとうございます。学部からの入学がいかに大変か、本当に尊敬します。色々な声を目にしましたが、アメリカ、特に大学卒の人たちは多様性を重んじ、他者を尊敬します。.
1
0
3
Beautiful work! Congrats Shawn!.
The VINE complex is a VPS9-domain GEF-containing SNX-BAR coat involved in endosomal sorting #biorxiv_cellbio.
1
0
3
I think this one of the best review for mechanistic insight of autophagy (especially yeast people). Great work! Also, thanks for citating my PhD work! Hahaha. .
Angelina's (@Ansa_Lina) and Martin's review summarising the "Mechanisms of Autophagy in Stress Response" .@ELS_CellBiology is now available for free download:.
1
1
3
The beautiful work, led by @MakinoShiho and Yoshinori Ohsumi, demonstrates selective mRNA degradation through autophagy, which definitely contributes to cellular remodeling in response to environmental cues. Congratulations!.
The Ohsumi lab shows yeast preferentially target specific mRNAs for autophagic degradation #autophagy @RIKEN_CPR.
0
0
1
Congrats to my previous labmate, Yui Tomioka, for her outstanding JCB paper, revealing the mechanim of selective degradation of the NPC complex via autophagy.
TORC1 inactivation stimulates autophagy of nucleoporin and nuclear pore complexes | Journal of Cell Biology | Rockefeller University Press
0
0
2
This is really cool.
A selective autophagy pathway for phase separated endocytic protein deposits #biorxiv_cellbio.
0
1
2
@KaoriYamada01 @minesoh 出典は忘れてしまいましたが、古いバージョンに近いレシピだと思います。精製が甘いポリクロには劇的な改善が見られ、名前と共にラボで流行ってます。笑.
0
0
2
This study developed extremely pH- and hydrolase- resistant Green fluorescent protein, which is pretty useful in various ways.
Online now! a signal-retaining #autophagy indicator, localized to #mitochondria (mito-SRAI) is able to quantitatively measure mitophagy and was applied to screen for chemical inducers of mitophagy and in a #mouse model of #Parkinsons disease. @RIKEN_CBS
0
0
2
This is prety useful resource! The game-changer has just shown up.
Endogenous tagging + 3D live-cell imaging + mass-spec interactome = an open-source map of the human proteome. So excited for our first release, wonderful collaboration with @labs_mann @loicaroyer & many other colleagues. @OpenCellCZB
0
0
2
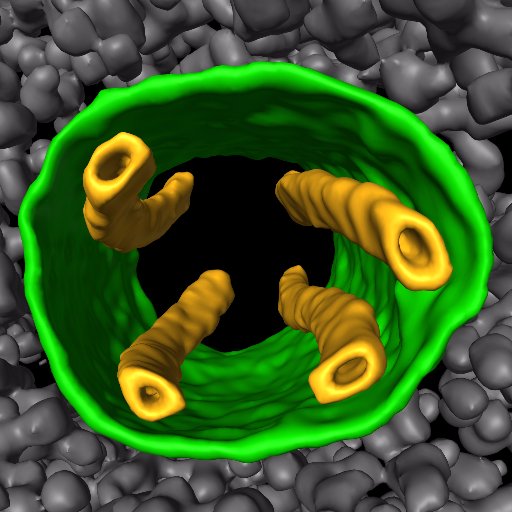
Congrats, Rick! Beautiful story!.
It's out! Twitterless Rick Hooy solves a #CryoET structure of clathrin-independent, adaptor protein complex that is hijacked during HIV infection. Thanks to Yui and Snow along with @DanTudorica3 for bringing this story together. Thread below: 1/9
0
0
1