Simon Gravel
@SFGravel
Followers
1,386
Following
92
Media
15
Statuses
394
Population genetics at McGill university. @SFGravel @ecoevo .social
Joined August 2012
Don't wanna be here?
Send us removal request.
Explore trending content on Musk Viewer
Eagles
• 230970 Tweets
Dick Cheney
• 192590 Tweets
Packers
• 130317 Tweets
ENGFA ACTRESS 100M
• 118238 Tweets
श्री गणेश
• 113163 Tweets
Ganesh Chaturthi
• 103865 Tweets
Ganesh Chaturthi
• 103865 Tweets
गणपति बप्पा
• 81210 Tweets
Saquon
• 76054 Tweets
Jalen
• 58603 Tweets
Barkley
• 44004 Tweets
भगवान गणेश
• 41183 Tweets
#二度と撮れない画像を貼れ
• 34726 Tweets
Jordan Love
• 32308 Tweets
#GanpatiBappaMorya
• 28779 Tweets
Party Love AndaLookkaew
• 26724 Tweets
JustinOn NCAA100Kickoff
• 25401 Tweets
ビッグラン
• 15478 Tweets
イコラブ
• 12766 Tweets
#学マスデビュー初心_名古屋
• 11232 Tweets
Nikocado Avocado
• 10555 Tweets
Last Seen Profiles
Many papers reported genetic evidence for 'ghost' archaic admixture in early human history. With new WGS data from Nama (Khoe-San) & other African individuals and parameterized modelling, we (
@apragsdale
@HennLab
et al) reach different conclusions.
7
81
214
A figure in the All of Us paper was criticized on many fronts, including the use of a dimension reduction technique (UMAP). Some people concluded that UMAP is not appropriate for representing genetic ancestry. I disagree.
@jkpritch
@ras_nielsen
@dgmacarthur
1/n
5
26
142
Lots of interesting recent methods & results on intricate models of human genetic history. We're having a mini-symposium next week May 6-7 to follow up on discussions at
#probgen21
.
Free registration:
4
56
108
Submission rejected at
@biorxivpreprint
because it included a French version of the abstract (preprint, in English, is on French Canadian genetic history).
Only English allowed.
Are you afraid I'll sneak in offensive material in the French abstract?
6
17
99
It may be time to retire or resequence some pioneering sequencing datasets. (With
@LukeAnderTroc
)
4
35
73
Coalescent theory allows for powerful simulations (eg
@jeromekelleher
’s msprime), but is biased for large sample sizes. Student D. Nelson et al. found a fix for large biases in IBD and ancestry distributions, expanding on an idea of Bhaskar et al 2014.
3
30
73
If you like UMAP, you'll love this preprint allowing for much easier interpretation of this structure you've been staring at.
If you hate UMAP, you'll love that this preprint formalizes a lot of hand-wavy interpretations of UMAP plots. (Ok, maybe not love it. Appreciate?)
1
18
60
Mini-workshop next week, please RT!
------------
Adaptation in structured populations
Thursday, June 29
------------
Featuring
@apragsdale
@AmyAGoldberg
@jblanc12
@QuintanaMurci
@nanditagarud
.
Free registration:
Program overview:
1
57
57
Our study on the history, genetics, and mathematics of the Great Migration of African-Americans
@PLOSGenetics
3
30
50
@apragsdale
's paper is out in Nature!
Coverage by
@carlzimmer
@NYTScience
:
And a perspective by
@DrEleanorScerri
:
Here's my original explainer:
Many papers reported genetic evidence for 'ghost' archaic admixture in early human history. With new WGS data from Nama (Khoe-San) & other African individuals and parameterized modelling, we (
@apragsdale
@HennLab
et al) reach different conclusions.
7
81
214
6
19
42
Preprint by postdoc
@i_krukov
: .
Most methods for modelling distributions of allele frequencies rely on either weak selection or small sample size assumptions. Ivan shows how to jointly handle large samples and strong selection.
1
18
41
Also, Dominic Nelson's Wright Fisher extension to
@jeromekelleher
's msprime is just out in PLoS Genetics
. Stay tuned for more large scale popgen simulations.
1
19
40
@LukeAnderTroc
explored the appearance of population structure in Quebec over centuries in using thousands of genomes and millions of genealogical records! . Story involves rivers, mountains, Royal hunting grounds, and asteroids.
3
10
31
Congrats to
@LukeAnderTroc
who successfully defended his PhD thesis today! Here he discusses the impact of an ancient meteorite on genetic diversity in Québec.
3
2
32
French version of this tweet:
Notre soumission a été refusée chez
@biorxivpreprint
à cause d'un résumé en français, cibole. Y veulent yenk de l'anglais, baswell. Chu t'en tabarnouche.
1
1
30
@apragsdale
heroically validated this model against multiple lines of genetic evidence that had been previously used to argue for archaic admixture, including LD statistics, the conditional SFS, and cross-coalescence rates. The model does well for all.
1
1
25
Making mistakes in genetic simulations is all too easy. Some suggestions for users, developers, and the entire community to make research results more robust.
Do you use msprime? You'll want to check out this commentary in our latest issue "Lessons Learned from Bugs in Models of Human History"
@apragsdale
@SFGravel
@jeromekelleher
1
7
14
0
7
24
Congrats to postdoc
@apragsdale
who will be joining the faculty at UW Madison
@UWiBio
! Also, congrats to
@UWiBio
for hiring such an outstanding scholar! Working with Aaron has been a blast!
1
0
22
Open rank search at
@mcgillu
in genomic medicine: .
A rare opportunity with enough resources to build computational + experimental programs!
0
12
19
UMAP is appealing because it can reveal patterns in the data that would not have been obvious otherwise. By contrast with PCA or admixture, it can reveal multiple levels of discrete and continuous population structure in one plot. See
@adp_diaz
’s papers & thesis on the topic. 5/n
3
5
19
Ahead of discussions+training next week at
@McGillGenome
about equity in genomics research training, I wanted to share these useful
@UCSF
guides for trainees and profs on talking about race and inequality in science.
0
5
17
@ProfLikeSubst
Given that some PIs actively discourage trainees from having kids and claim that they are career-killers, it's fine to have some researchers point out that having kids is ok and can even be nice. Obviously, shouldn't be directions or unsolicited advice to their trainees (please!)
1
0
15
Lots of other cool finalists at this year's
#ScienceExposed
data viz competition, but
@adp_diaz
's UMAP figure is the only non-imaging one -- support your fellow data nerd and vote :)
1
3
14
Tenure-track position at McGill in statistical genetics:
http://t.co/Rw9uxBMcu7
It's an awesome place to do stat/popgen research!
0
50
14
Our latest preprint: The Great Migration and African-American genomic diversity
http://t.co/Jnw1PuEQsf.
Congrats to postdoc Soheil Baharian!
0
15
12
Abstract deadline now February 13 for Recomb-Genetics (May 4, 2019, Washington DC). Keynote speakers Sharon Browning and Josh Akey, ahead of
@recomb2019
featuring
@cdbustamante
,
@Alfons_Valencia
et al.
Expect haplotypes and ancient history!
Please RT :)
0
10
10
I join
@LukeAnderTroc
in thanking our co-authors
including Dominic Nelson, Shadi Zabad,
@adp_diaz
@jeromekelleher
, Ivan Krukov,
@benjeffery
@genotepes
,
the teams at
@_CARTaGENE_
and
@BALSAC_UQAC
, and (43,000 times) the generous participants at
@_CARTaGENE_
1
0
9
Those of you at
#SMBE2019
should check out
@apragsdale
's talk on Wednesday! Heroic work including theory, software, and applications.
0
2
9
Ten days left to submit abstracts for Recomb-Genetics (May 4, 2019, Washington DC).
Guest speakers Sharon Browning and Josh Akey, just ahead of
@recomb2019
featuring
@cdbustamante
,
@Alfons_Valencia
et al.
Expect haplotypes and ancient history!
0
7
9
Beau travail de
@LukeAnderTroc
pour visualiser
#covid
avec les données de
@OpenGovON
. Pourrait-on avoir les mêmes données à
@sante_qc
?
I wrote an
#opensource
script that automatically downloads and plots the latest
#COVID19
data from
#Ontario
. Scripts are available here :
#DataVisualization
#DataScience
#covid19Canada
#ontarioshutdown
1
13
42
1
2
9
@leland_mcinnes
@adp_diaz
We ran the
@1000genomes
project data without PC filtering, and it looked very good
0
2
9
Thanks
@HennLab
! It was great to catch up! I may want to use the top right photo as cover art for
@LukeAnderTroc
's next manuscript...
0
0
7
@kzshabazz
@monicaMedHist
@SandyDarity
Ancestry+demography (not race) are important to understand genetic diversity
2
8
8
@LukeAnderTroc
describes batch effects in the 1000G project (and their consequences) .
You may want to check your favourite imputed SNPs against his list of batchy SNPS,
,
Also check out those of
@FabriMafe
!
1
5
8
@michaelhoffman
I should have reserved that domain! My favorite gravel sign so far still gets regular use on my office door:
1
0
7
@adp_diaz
' approach using
@leland_mcinnes
and Healy's UMAP applied to GnomAD data
(UMAP:
Alex's paper )
0
1
6
Therefore genetics research should not stop at arbitrary boundaries set by a racist society. 3/3
@monicaMedHist
@kzshabazz
@SandyDarity
5
6
6
With talks by
@apragsdale
@sr_sankararaman
&Erin Molloy, Paul Verdu,
@HennLab
@morestrada
, and
@QuintanaMurci
0
0
6
Marketing emails from ASHG vendors are off the chart this year. Both in terms of volume and spamminess. Assuming some email list leaked?
@GeneticsSociety
, any chance you can rein in vendors?
0
0
6
@sbmontgom
@marcus_NJ
@popgengoogling
@AwadallaLab
@ArmandeAngHoule
@hgibling
@KimSkead
As a French Canadian, I would never joke about poutine. It's clearly an outsider.
0
0
6
PhD and postdoc positions at McGill University. Involves genetics, math, history, biology, computing, and cool data.
http://t.co/tbhCMhCMq4
0
12
5
We were also thinking of funding this project through the sale of art prints, but then I'm not sure we can use
@uk_biobank
data for commercial purposes :)
0
0
5
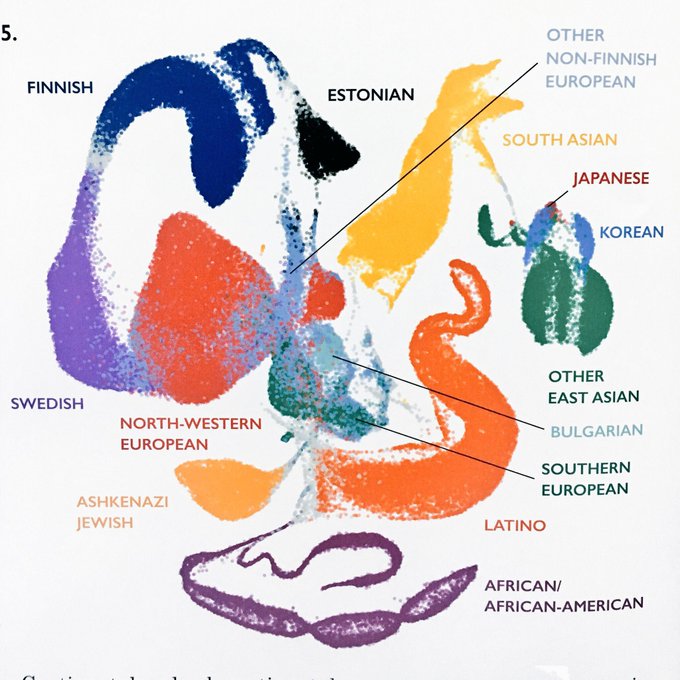
The UMAP method was developed by
@leland_mcinnes
and John Healy for visualizing high-dimensional data (). Each dot is an individual in the UK biobank, coloured by self-identified ancestry categories ( )
1
0
6
@adamauton
@rdhernand
@LukeAnderTroc
Someone should analyze the AFS (acronym frequency spectrum) for the 1000GP/TGP/kGP
0
1
6
Tests génétiques: attention aux tests bidon! Aucun test d’origine génétique ne donne de statut légal! via
@lp_lapresse
0
3
5
This is starting in 30 minutes!
Lots of interesting recent methods & results on intricate models of human genetic history. We're having a mini-symposium next week May 6-7 to follow up on discussions at
#probgen21
.
Free registration:
4
56
108
0
2
5
@lu_sichu
Race is a socially defined label that is correlated with ancestry. So if you label individuals by race on a dimensionally reduced dataset, you will see patterns. But umap will also show variation, say, among French Canadians living across Quebec.
1
0
4
@rdhernand
Or it's all coding but French is more robust to errors!
So CIHR could check whether scores for applications in French have lower variance across reviewers :)
2
0
5