KinesinMotors
@KinesinM
Followers
164
Following
2K
Statuses
2K
Motors, Microtubules, Mechanobiology My views only Now at https://t.co/noMXfHqDym
NC, USA
Joined November 2019
Wonderful! Can't wait to download and read it - keep going!!
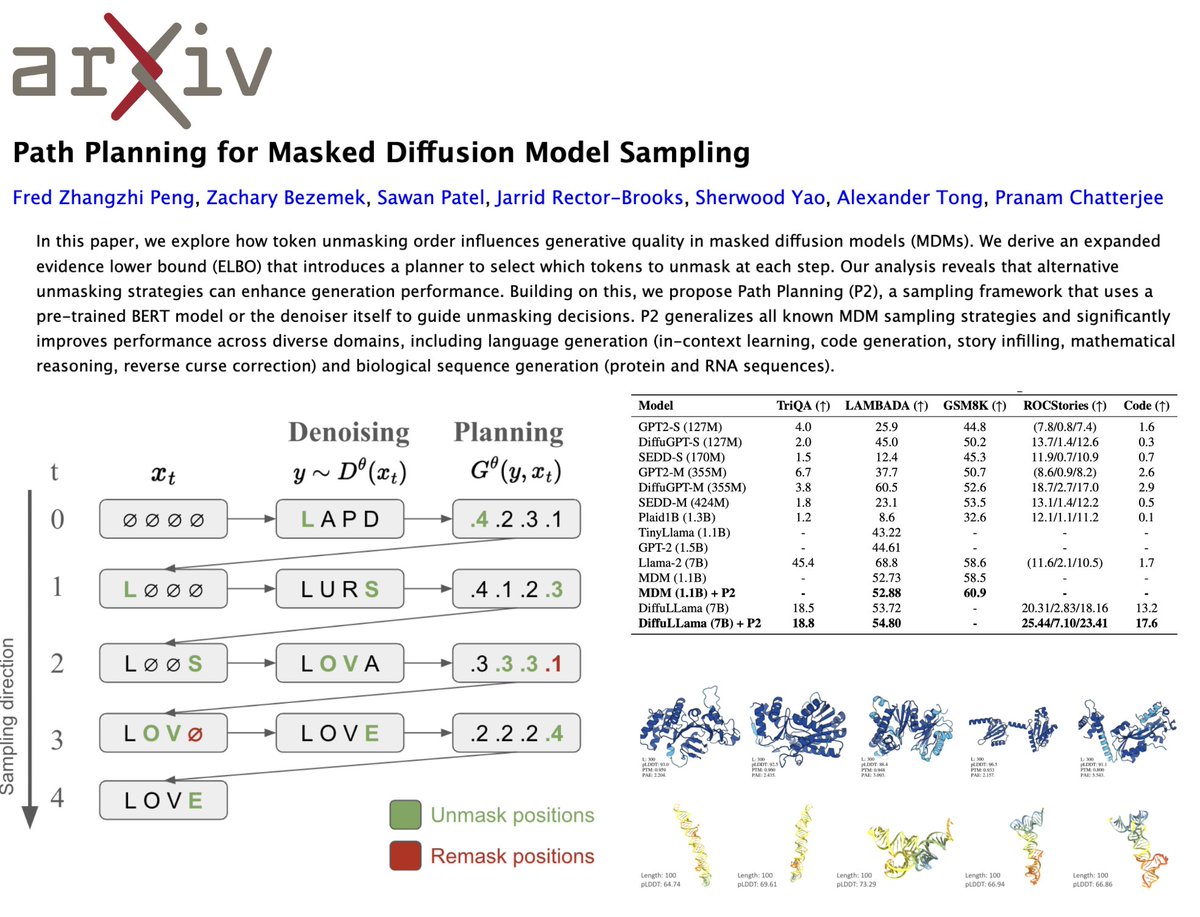
It's tough times for science. 🥺 But we have to keep innovating to fight another day, and today I'm so proud to share @pengzhangzhi1's new, groundbreaking sampling algorithm for generative language models, Path Planning (P2). 🌟 📜: 💻: In the appendix!! 👆 Masked discrete diffusion models (MDMs) have significantly outpaced autoregressive models in generation quality, ESPECIALLY for biological sequences which are inherently bidirectional! 👉👈 We've seen it work for both wild-type and modified peptides as well as for membrane protein design! 🥳 Still, MDMs struggle with inefficient token unmasking, which can lead to suboptimal outputs. 🫤 P2 fixes this by optimizing the order in which tokens are revealed, using either a pre-trained BERT model or the denoiser itself to make smarter, context-aware unmasking decisions. This eliminates randomness, leading to smoother, more accurate generations across tasks!! 😎 P2 achieves state-of-the-art performance across multiple domains! 🔥 In math reasoning, a 1B MDM model outperforms a 7B LLaMA. In code generation, P2 crushes autoregressive models on functional correctness. For biological sequences, P2 + DPLM produces the highest-quality proteins and even RNA sequences that fold better than real biological sequences! This is sampling done right. 🦾 I'm always so impressed with how brilliant @pengzhangzhi1 is (remember our PTM-Mamba paper? It's coming out soon btw! 🐍), but also with how he works with others! 🤝 This is a result of a strong partnership with @DukeU math faculty Zachary Bezemek, as well as a close collaboration with @sawanp_813 and @sherwoodyao for RNA generation, and @jarridrb and @AlexanderTong7 for theoretical insights! 💡 Take a read of our paper, and with the sampling code in the appendix, it's super easy to add to your favorite discrete diffusion model for high-quality generation!! 🫧
0
0
0
RT @SiaraRouzer: A Temporary Restraining Order has been Granted Against NIH IDC Cuts, until a hearing at 10am on February 21. https://t.…
0
68
0
Wondeful - thanks for the life line, NC AG Jackson !!
BREAKING: Attorney General @JeffJacksonNC just won a temporary restraining order preventing the NIH from unlawfully stripping funds that support cutting-edge medical and public health research at universities and research institutions across the country.
0
0
1
RT @ItaiYanai: Science is key to the US's success but this government's intention to dramatically reduce the part of the federal grants tha…
0
48
0
Thank you, @JeffJacksonNC ! We ask you now to block and declare illegal the NIH Director's order reducing NIH grant indirect costs to 15% for all institutions on Feb 10, 2025 and going forward. This order has immediate and disastrous consequences for scientific research in NC.
This morning, a judge blocked the federal government from unlawfully sharing Americans’ personal and financial data with DOGE. Attorney General @JeffJacksonNC sued last night and helped win the order, protecting North Carolinians’ private data.
0
0
2
RT @pranamanam: As you can probably tell from our work, we're now all in on diffusion and flow matching models for protein/peptide sequence…
0
30
0
@JeffJacksonNC Thank you! Now please take action to freeze the anti-DEI EO issued on Jan 20, 2025 - this is causing NIH & NSF to halt grant funding & review, costing millions of $$ in research time and effort to NC universities
0
0
4
🥂🌹🎆
Okay, another VERY exciting paper drop! 🎉📜 So you guys may know the story of how @GametoGen was started? 🐣 My part of the story begins with a visit to @geochurch at @harvardmed, with a (perhaps naive) goal: to make an oocyte from a stem cell. 🧫 With @GametoGen's support, I started working on it together with @krammecc, and later with @mpsmela1. 5 years later, after many rejections and sleepless nights of experiments/analysis, our paper describing oogonia differentiation from iPSCs is finally published in @emboreports! 🎉 📜: Ever since the generation of iPSCs from somatic cells, stem cell differentiation to different cell types has been a major challenge. Some of the toughest cells to make are gametes (oocytes and sperm) -- they have crazy methylation dynamics, they are haploid, and they can obviously produce offspring !👶 The search space for the right cocktail of molecules and transcription factors (TFs) to make these is complicated and vast -- a perfect problem for an algorithm! 💻 As a part of our STAMPScreen pipeline (, we developed an adaptive PageRank-based algorithm that could rank "central" transcription factors in the gene regulatory networks of differentiating cells. 📊 After screening the top 46 TFs ID'ed for oocyte production in iPSCs (while also pushing them through primordial germ cell-like cell (hPGCLC) formation), we identified 3 that both independently and together (D3), drove DDX4+ oogonia formation, as well as 3 that drove improved hPGCLC formation! 🤩 We have since showed that cells we engineered with these factors, termed induced oogonia-like cells (iOLCs), can be generated in just FOUR DAYS (not like the previous FOUR MONTH protocol) and exhibit late-stage oogonia-like gene expression, epigenetics, and protein markers. 🧫 They are defined by the expression of mitotic oogonia markers (DDX4, DAZL, and KI67) and the absence of earlier primordial germ cell and pluripotency markers (OCT4 and SOX17). Remarkably, these iOLCs are capable of feeder-free expansion culture while retaining their oogonia-like identity post-sorting, making this a super rapid, scalable, and robust system to study human germline development! 💨 Honestly, this was only possible via my incredible partnership with @krammecc (now the CSO at @GametoGen!) and our brilliant collaboration with @mpsmela1, as well as with @Patrick11F, my @DukeU undergrad Shrey + the entire team! 🙌 It was just sheer perseverance for years as we CONSTANTLY got rejected by journals and had to keep adding new data and analysis (I thought it would never end). We were also so lucky to have the guidance of the Shioda Lab at @MassGenBrigham and the team at the @wyssinstitute through the process! 🤝Just so many great scientists who put their everything into this! 🫶 I know a lot of you know me as a peptide/protein designer (or probably just as the sequence-only guy), but this shows you how impactful good algorithms can be! Who would have guessed a graph theory algorithm could be used to make human oogonia?! 🙃 I'm just really excited for my lab to continue work in this space and make an impact in reproductive medicine with our new generative models! 💻 I hope you can take a read of our paper and let us know your thoughts! 🙏
0
0
0
NIH panel cancellations appear to be related to the announcement below
I am told that @NIH grant review panel dates (these always are scheduled 3-6mo ahead) are getting cancelled left and right (no pun). Does anyone know what this is about? @DrJBhattacharya @RobertKennedyJr
0
0
1
RT @PracheeAC: It was spring 2020. I was a professor and had recently learned I’d successfully been awarded early tenure. At the tail end o…
0
30
0
All the best, Ruth !!
Today is retirement day for our amazing lab manager, Ruth Montague. Ruth provided years of first-class service to Dan Kiehart's lab @DukeBiology and (since 2011) our lab @Duke_PCB. Enjoy more time with the horses Ruth! You deserve it.
0
0
0
RT @pranamanam: So excited to host the 2nd GEM Workshop at ICLR 2025! 🎉 We have amazing speakers/panelists 🧑🔬, money for new AI+Experiment…
0
5
0
RT @GladfelterLab: Excited to share our recent work on the power of RNA to encode physical information and how this can be embedded in the…
0
31
0
RT @Duke_PCB: Big congratulations to our colleague, Dr. Zhang (ZZ) @zz_zhaozhang, a recipient of the @sontagfdn Distinguished Scientist Awa…
0
1
0
RT @pranamanam: 🚨 Current graduate students! If you're interested in developing and leveraging generative language models for therapeutics…
0
7
0
RT @GladfelterLab: Excited to share the latest work from postdocs @turiyaaa and Zack! We thought about how multinucleate cells are able to…
0
19
0
Such a wonderful advance 🌹
Surreal! 🤩 Along with @martinvars and @DinaRadenkovic, we co-founded @GametoGen in 2020 with just a silly graph theory algorithm I designed to predict TFs that could differentiate ovarian cells from iPSCs. 💻 Now, little Mia is here with the technology that has grown out of that work. 🐣 It's completely due to the scientific brilliance and visionary leadership of @DinaRadenkovic, @krammecc, and the entire team at the company -- I'm so proud of them!! 🥰 And I'm super excited for this to fundamentally shift the field of reproductive health!! ⚕️
0
0
0
RT @FoxLabDuke: #CellBio2024 : check out Fox lab postdoc @archan24 's poster tomorrow (Tuesday) to find out how cells balance mitochondrial…
0
8
0