Pranam Chatterjee
@pranamanam
Followers
3K
Following
1K
Media
44
Statuses
541
Designing proteins to program biology! 🧬💻🧫 Assistant Professor at @DukeU | Co-Founder @GametoGen and @UbiquiTxINC | Formerly @MIT '16 '18 '20 and @harvardmed
Cambridge, MA
Joined August 2015
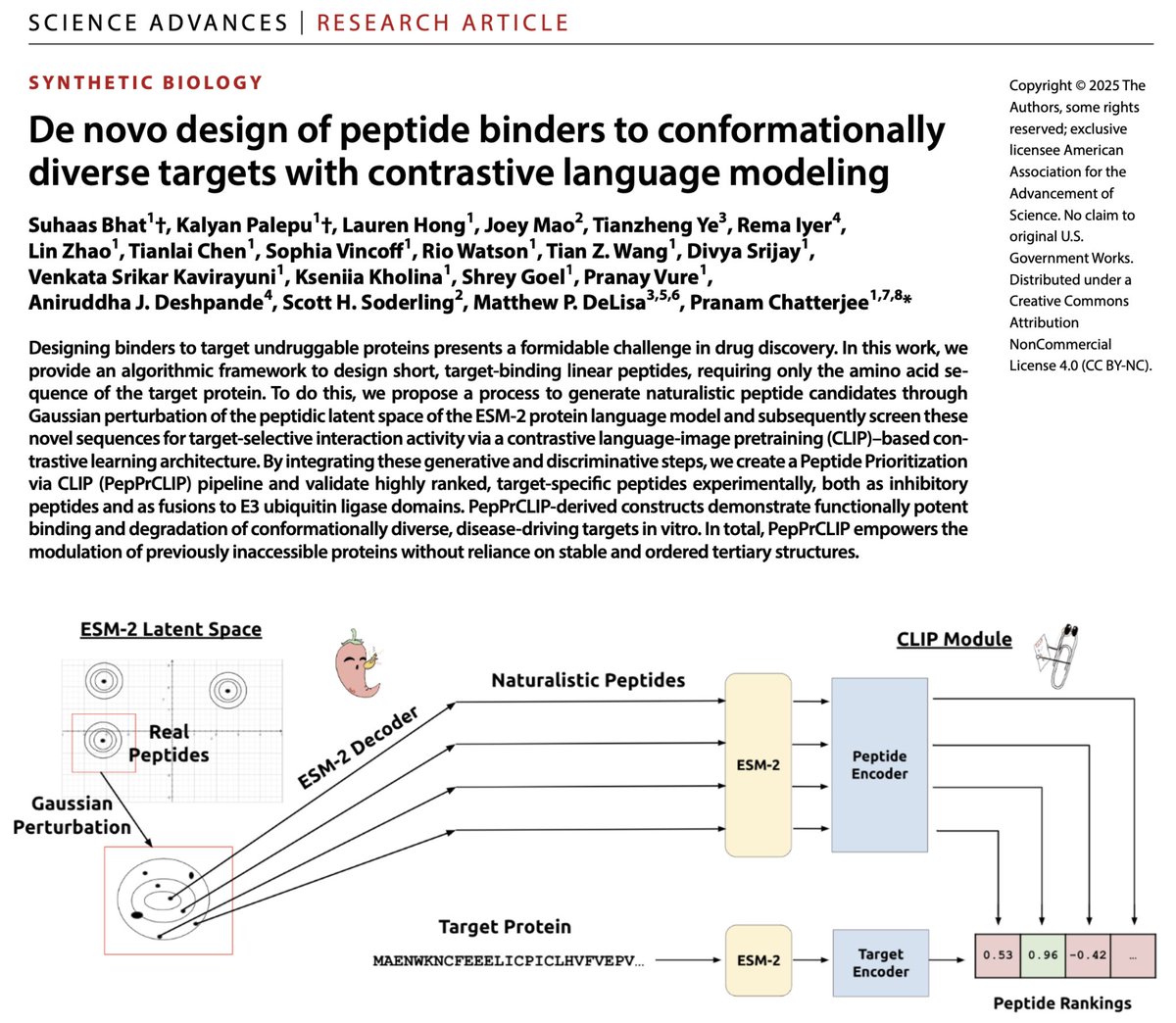
I'm usually not too emotional about paper acceptances, but this one justifies it. 🥹 It doesn't feel real, but PepPrCLIP is now published at @ScienceAdvances! Let me tell you its story. 📕. 📜: 💻: PepPrCLIP (🌶️📎) began as
11
55
376
A beautiful thread from @rohitsingh8080!! 🧵 It really captures my feelings/frustration about current "trends" in protein design. As Rohit mentioned, the aftermath of sequencing the human genome has been a huge huge disappointment. Genomics has really not lived up to the hype,.
Let me tell you a story. It'll end up at the current tech-bio and protein design scene. But the story starts about 25 years earlier. Did you know that, commercially, the human genome project precipitated the end and not the start of a genomics boom? 1/.
17
38
296
Super excited to share our new protein language model: FusOn-pLM, developed by my wonderful first-year PhD student @SophieVincoff and her team of amazing undergrads!! 🎉 . Sure, we know AlphaFold3/RFDiffusion and other structure prediction/design methods are great -- but what
8
38
178
Hi everyone! Excited to share that I will join the amazing faculty at @DukeU as an Assistant Professor of Biomedical Engineering in July! We will design therapeutic proteins for genome editing, proteome editing, and ovarian cell engineering! 💻→🧬→🧫→🏥.
24
5
157
#NeurIPS2023 was so much fun! My favorite talk from the workshop has to be from @PrescientDesign! Similar to @OpenAI's Consistency Model formalism, they sample a score-based data manifold with one step denoising for antibody design! #GenerativeAI.
3
21
135
This is the beauty of collaborative science! 🧪 Over the summer, my good friend @htkratochvil and I were thinking about how we could generate membrane proteins (her lab's expertise) with our lab's generative pLMs. Obviously, structure-based methods aren't great as membrane
4
22
132
An exciting @biorxivpreprint update on our recent PepPrCLIP model!🌶️📎 Hopefully, you all remember PepPrCLIP, where we apply Gaussian perturbations to the peptidic latent space of ESM-2 to de novo generate naturalistic peptides, and then input these new peptide sequences into a
0
21
128
Interesting new pLM (AA-0) from @Ginkgo! It definitely seems they have found, as we've discovered many times over, that ESM-2-650M is the ideal baseline pLM, but Ginkgo does make some notable "enhancements" with their model. Architecturally, nothing different, but the training
3
14
124
Okay, this is SO COOL from @Mila_Quebec: Aaren, which leverages a parallel prefix scan algorithm, can be trained in parallel (Transformers) but only requires constant memory (RNNs)! 🤩 It's kind of similar to RWKV and Linear Attention, but unlike those methods, Aaren exactly
0
20
110
Excited to share our newest Cas9 out in @NatureComms! 🥳 We recombine our Sc++ enzyme (NNG) with the PID of @BKleinstiver's SpRY (NRN) to generate a PAM-flexible chimeric Cas9 = SpRYc! 🌶️ SpRYc can edit at diverse NNN loci -- take it out for a spin! 🧫
3
27
106
Exciting news from the lab! 👀 We have updated PepMLM on the @arxiv with new computational and experimental benchmarking results! 🥳 . Just as a reminder: we trained PepMLM via a novel span masking strategy that positions peptide binder sequences at the C-terminus of their target
5
21
98
Whoa!! @EvoscaleAI just came out with a potentially useful upgrade to our favorite ESM-2 pLM with three new ESM C representation models: 300M, 600M, and 6B! 😳 Here, I'll do three semi-analyses based on the release: model modifications, "evaluations", and license. Would be happy.
Introducing ESM Cambrian. Unsupervised learning can invert biology at scale to reveal the hidden structure of the natural world. We’ve scaled up compute and data to train a new generation of protein language models. ESM C defines a new state of the art for protein
3
9
94
🧵 (1/4) As a brand-new PI, I'm psyched to present my group's first preprint! Here, we apply @OpenAI's CLIP architecture to design target-specific peptides, fuse them to E3 ubiquitin ligases, and degrade pathogenic proteins in a pipeline called Cut&CLIP. ✂️
7
11
90
Just getting around to posting about this beautiful paper from @DreamFoldAI!! Learning a joint representation based on SOTA sequence/structure embeddings and decoding the joint representations of the inputs into SE(3) vector fields is very clever -- my hunch is that the ESM-2
We are thrilled to announce FoldFlow-2: our new SOTA protein structure generative model. w/ @Guillaume_hu James V @FatrasKilian Eric TL @PabloLemosP @riashatislam @ChengHaoLiu1 @jarridrb @tara_aksa @mmbronstein @AlexanderTong7 @bose_joey. 🪄 Blog post:
1
18
90
Surreal! 🤩 Along with @martinvars and @DinaRadenkovic, we co-founded @GametoGen in 2020 with just a silly graph theory algorithm I designed to predict TFs that could differentiate ovarian cells from iPSCs. 💻 Now, little Mia is here with the technology that has grown out of that.
(1/) We are thrilled to announce a medical milestone–the world’s first live birth using our product, Fertilo, that matures eggs outside the body.
8
7
84
I am 100% on the side of the DiffDock team on this one, even as a someone who believes almost all structure-based docking models are artifactual. Some things to note:. -The more "standard" docking pipelines that were compared against DiffDock on head-to-head tasks are largely.
You may have seen a recent pre-print [1] from Jain et al. with strongly worded claims against the experimental results in our DiffDock paper [2]. We initially declined to respond as we saw that this preprint contained falsehoods, misleading comparisons, seemingly deliberate.
5
8
79
Today, I "officially" begin my faculty position at @DukeU! 🥳 Super excited to welcome my wonderful team of postdocs, PhD/Masters students, and undergrads from around the globe to our little but mighty lab in Durham ― let's change the world together! 💪🏾
5
7
76
SaLT&PepPr is published in @CommsBio! 🥳 Here, we fine-tune the ESM-2 pLM to identify peptidic binding sites on target-interacting partner sequences. We fuse these "guide" peptides to E3 ubiquitin ligases to degrade disease-causing proteins! 💻➡️🧫 (1/n)
5
15
74
A great day for our field, and very well deserved!!! 🏅 The work done at @UWproteindesign and @GoogleDeepMind, and of course that of @geoffreyhinton @HopfieldJohn, and the RNAi/miRNA community, especially at @UMassChan, have surely changed my life, and have inspired so many.
BREAKING NEWS.The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”
1
9
73
How can AI help us cure rare diseases? In the Aug/Sep @WorkingAtDuke magazine, I talk about my lab @DukeUBME and how we’re training generative language models to help us design drugs to target complex proteins. Keep an eye out for the magazine in your mailbox! @dukeresearch
1
8
72
Such a beautiful study from our friends in @rohitsingh8080’s lab here at @DukeU! An autoencoder on top of ESM-2 for fixed dimensionality is brilliant, and perfectly suited for this problem, one that has plagued delivery and other drug design applications. 🌟 I can imagine so many.
De novo protein design is great, but nature has millions of proteins- why not repurpose them? . Introducing Raygun, a new approach to protein design. It allows you to miniaturize, magnify or modify any protein. We synthesized miniaturized variants of eGFP and mCherry! 1/
2
16
70
So many congratulations on to my very brilliant friend @MartinPacesa on this really amazing achievement with BindCraft!! It's a very "crafty" use of AF2-Multimer weights and super principled validation and filtering with AF2 metrics and Rosetta physics-based scoring. metrics. 🌟.
Have you ever wanted to design protein binders with ease? Today we present 𝑩𝒊𝒏𝒅𝑪𝒓𝒂𝒇𝒕, a user-friendly and open-source pipeline that allows to anyone to create protein binders de novo with high experimental success rates. @befcorreia @sokrypton.
1
8
68
Try out @pengzhangzhi1's PyTorch implementation of ESM-2 with FlashAttention! 🔦 So easy to use that I even I 😵 was able to drop it into some of my own code and get it to work in seconds. It's always so nice to have Fred making life easier for us in the lab!! 🫶.
ESM2 is our go-to pLM. However, running, fine-tuning, or training a model with millions of parameters can be costly. We reimplemented ESM2 with FlashAttention, achieving up to 60% memory savings and 70% faster inference! 🚀Check it out on github: (1/n)
1
7
66
Hey guys, I think I may have shared too much about my feelings (as usual) about protein design in the last post! 😅 I definitely don't mean to put down any line of research, and I think all scientific exploration is important -- that's why I love being a scientist and engineer!.
Interested in how #LLMs are used in #health? I'm so excited to have @pranamanam giving the first virtual seminar of the semester in the @DukeEngineering Computational Health Seminar Series. Anyone can join in remotely to learn more about his innovative approach!
7
2
64
Okay, another VERY exciting paper drop! 🎉📜 So you guys may know the story of how @GametoGen was started? 🐣 My part of the story begins with a visit to @geochurch at @harvardmed, with a (perhaps naive) goal: to make an oocyte from a stem cell. 🧫 With @GametoGen's support, I
0
11
65
Hi everyone! I am very excited to announce that our broad, efficient, and specific enzyme, Sc++, has been published in @NatureBiotech! 🤩 An incredible collaboration with @njakimo @JooyoungLee10 @SontheimerLab @medialab @UMassMedRTI 🎉🎊 Check it out! 🔽 (1/5).
3
15
60
This is such cool work published in @NaturePhysics! 🤩 An N-terminal unstructured and flexible glycine–serine peptide tag can accelerate nuclear import rates for stiff protein cargos by increasing their deformability. Definitely useful for my experimentalists to deliver our
0
4
60
Well here it is: ESM3!! 🌟 The training regime is a bit different than your classic ESM-2/BERT model: you can start with a fully masked sequence and iteratively unmask. Super similar to our PepMLM model with ESM-2, which showed significant promise at full masking and unmasking.
We have trained ESM3 and we're excited to introduce EvolutionaryScale. ESM3 is a generative language model for programming biology. In experiments, we found ESM3 can simulate 500M years of evolution to generate new fluorescent proteins. Read more:
1
9
56
I'm so excited to co-host the @gembioworkshop at #ICLR2024 in Vienna!! 🥳To both my computational and experimental friends, please reach out if you are interested in submitting a paper or serving as a reviewer -- it should be an incredible, one-of-a-kind meeting! 🌟.
Announcing the Generative and Experimental Perspectives for Biomolecular Design workshop at #iclr2024!.We hope to bring together researchers in ML and experimental biology to accelerate progress on real-world applications. Website: .Paper deadline: Feb 3
3
11
51
You guys know how much of a sequence-first mindset I have! 😅 Based on experimental evidence from my own research/lab and our papers, I really do believe that pLM-based generative algorithms can have an impact where structure-based generators fail (which for proteins we work on,.
🚨Discrete diffusion models are the next big thing. But guess what? RLHF-style fine-tuning and guidance for them aren’t the answer! ❌. What does work? 🚀The simulation-free DDPP objective. Check out our latest work for all the details. 👇 a 🧵. Arxiv:
0
9
52
A little late, but super excited to share a great collaborative work by our team at Duke, my postdoctoral lab of @geochurch, and my company @GametoGen to engineer a functioning human ovarian follicle from iPSCs using our STAMPScreen method! 🧫
1
7
49
After almost three years of painstaking effort, today, I'm excited to present my team's amazing work on specifying human oogonia via combinatorial transcription factor induction in iPSCs! 🥳 An incredible collaboration with my good friend @krammecc!
2
4
41
Beautiful stuff @AlexanderTong7 @mmbronstein and team!! MFM enables generative models to accurately match vector fields on data manifolds -- so many cool applications: LiDAR navigation, unpaired image translation, single-cell RNA trajectory inference. such impressive work! 😊.
If data lives on a manifold, how do we design meaningful interpolations between marginals? We present Metric Flow Matching (MFM)…. @PPotaptchik @TeoReu @leoeleoleo1 @AlexanderTong7 @mmbronstein @bose_joey @Francesco_dgv . 🔗Dive in here: 🧵 (1/12)
1
11
43
So excited to host the 2nd GEM Workshop at ICLR 2025! 🎉 We have amazing speakers/panelists 🧑🔬, money for new AI+Experiment collabs 🤝🤑, and we're partnering with @NatureBiotech to get the best papers into review! 📜 Definitely submit your new work and see you in Singapore!! 🇸🇬.
We're BACK! 🧬 The 2nd GEM Workshop at @iclr_conf 2025 is ON!! 🎉 Exciting speakers, exp-comp matchmaking + seed grants, and a partnership with @NatureBiotech! 🌟 Submit your paper and join us in Singapore! 🇸🇬. Website: Papers Due: February 3rd, 2025 📜.
0
5
43
🚨 Current graduate students! If you're interested in developing and leveraging generative language models for therapeutics design, please apply to the @FutureHouseSF postdoctoral fellowship and indicate my lab as an option! 😃 $125k salary and access to all of @FutureHouseSF's.
FutureHouse is launching an independent postdoctoral fellowship program for exceptional researchers who want to apply our automated science tools to specific problems in biology and biochemistry, in collaboration with world-leading academic labs. --$125,000 annual stipend.
0
7
42
Excited to host the ByteDance AI4Science for a seminar on Protenix! 🤗 They’re absolutely one of my favorite AI teams in industry and have done some beautiful, open-source engineering for the AIxBio community! 💙.
Interested in Protenix, the open reproduction of AF3 from Bytedance AI4Science team? Curious to learn more? Luck for you! Join us for a special talk hosted by Chatterjee Lab ( on Thursday, January 16, at 10 AM EST. Zoom: (1/n).
0
4
41
Excited that my first company @GametoGen has just raised our Series B!!🧫 This is the amazing work of our incredible CEO @DinaRadenkovic and CSO @krammecc who have led the company to new heights since @DinaRadenkovic, @martinvars, @krammecc, and I decided to get this off the.
The AlleyWatch Startup Daily Funding Report for 5/29 ft:. STARTUPS: @GametoGen @authcaptives .FOUNDERS: @dinaradenkovic @martinvars @pranamanam @authcaptives .LEAD INVESTORS: @twosigmavc @firstmarkcap . . #NYCtech #startup #funding #VC.
4
3
41
A very cool use of graph-based diffusion for enzyme evolution! Also love the application to pAgo, which can be a very powerful programmable gene editing tool with improvements. #GenerativeAI #ProteinDesign.
0
9
39
Training a VAE-based representation model, performing diffusion on the latents to generate new aptamers, with SPR results show an average 3-fold Kd reduction? Probably needs a bit of refinement and validation, but so cool! 🫨 #GenBio.
0
4
38
Happy to share our early work on generating binding peptides conditioned ONLY on the target sequence! 🌟PepMLM masks cognate peptides at the end of target protein sequences, and tasks ESM-2 to fully reconstruct the binder region. 😷 #GenerativeAI
1
4
37
Hi all! We have released a preprint on our novel SaLT&PepPr language model to design peptide-guided degraders using PPI information. 🥳 An incredible effort from @garykbrixi and team at @DukeU, and our collaborators at @Cornell with @mattdelisa! 🤜🏾🤛🏾
1
6
34
Today is my happiest day as a scientist: my incredible undergrad @SabrinaKoseki was awarded the @NSF GRFP! 🍾 I've mentored Sabrina for 3+ years now, and her brilliance, curiosity, and diligence EPITOMIZES scientific excellence. I've been so lucky to have her as my student!😊
2
1
33
@NeurIPSConf Not surprised that the Spotlight paper is from a school like Groton. Let’s stop putting random pressures on high schoolers — this is really really getting out of hand. 😔 In a few years, a NeurIPS acceptance will be almost mandatory to get into places like MIT/Caltech/etc.
0
0
32
The Programmable Biology Lab is all set up at @DukeU — all in 1.5 days! 💪🏽 Shoutout to my amazing Harvard CS undergrads, Garyk and Suhaas, for helping make this happen. 🌟Time to design some programmable proteins! 💻➡️🧫
5
0
31
Nice! A great application of Mamba on concatenated, homologous protein sequences! 👍 Happy the authors mentioned our recent PTM-Mamba model, where we showed the application of Mamba to represent modified sequences! Overall, SSMs seem to have a found nice home in biology! 🧬.
🚀 Excited to introduce ProtMamba! Our novel protein language model designed to facilitate protein design.🔬📄 🔧 1/n
2
5
31
Super excited to announce the publication of our "AA" targeting Cas9, iSpyMac, in @NatureComms! 😍 Another incredible collaboration with @njakimo @JooyoungLee10 @SontheimerLab @medialab @UMassMedRTI 🎊Check it out 🔽 (1/5).
5
5
29
Check out our lab's perspective on generative design of therapeutic binders published in COBME: 💻🧫 Great work from my PhD students:.@LeoChan213 @lauren_hong11 @vivi_64_ @SophieVincoff Feel free to cite the review in your next manuscript! 😊#GenerativeAI.
1
9
30
We're so grateful to #EndAxD for believing in our binder-guided degrader technology, powered by #GenAI, to study and treat Alexander disease. 💻🧫 Thank you to @thomas_wagner and little Max for inspiring my lab to study, treat, and cure AxD! 🙏🏾
5
3
28
Remember @DeepMind's protein folding algorithm, AlphaFold2? I'm excited to share our team's early work on our own protein structure prediction pipeline! (1/3)
1
7
24
As the second week of my lab's existence comes to a close, I'm super grateful for the dedication and commitment of my students to our (quite lofty) goals. Yeah, we may not be the most well-known bunch, but we're about to do big things! 💪🏾 #FearlessFriday.
1
0
23
My last day together at the @medialab with my superhero team of undergrads, @SabrinaKoseki, Teodora, and Emma! So very proud to have watched you three become such brilliant young scientists, and so privileged to have mentored you as my first students! 🌟🥹
0
0
24
An integrated pLM trained on BOTH sequence and structure! 🤩 We're excited to leverage these embeddings in our lab -- the T5 architecture is super powerful for design! So many congrats to.@HeinzingerM and the @rostlab for another pioneering work! 👏
1
2
23
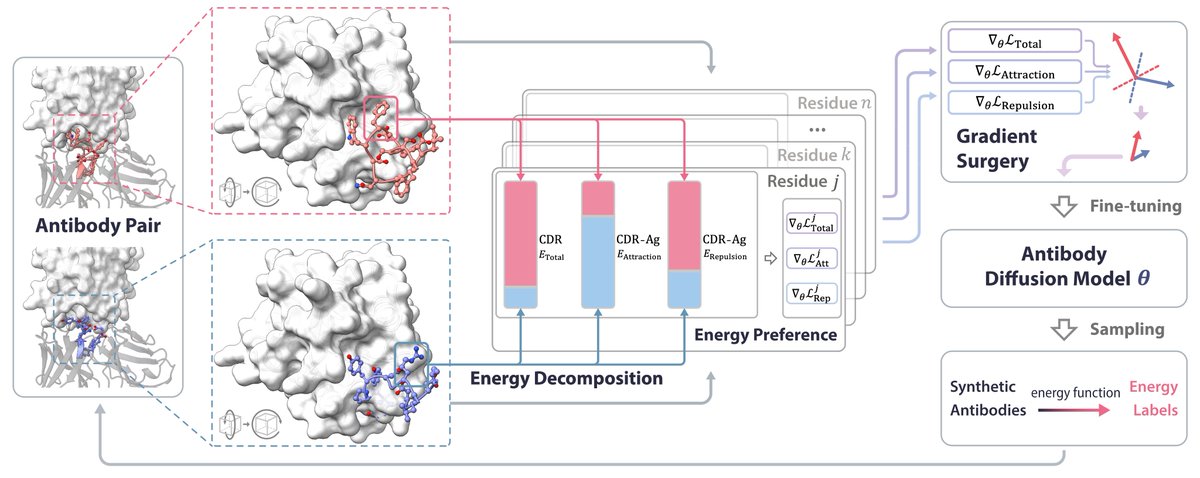
Residue energy-based preference optimization of antigen-specific antibodies. 🫨 A very interesting way to get around the lack of diverse paired structures for training!.
How to steer generative antibody design models to design antibodies satisfying user-desired preferences, such as rationality and functionality🤔? .We are thrilled to introduce AbDPO, a general framework for antibody design via energy-based preference optimization. The preprint,
0
1
23
Hi friends! Just a gentle reminder that our #ICLR2024 @gembioworkshop paper submission deadline is coming up in ONE week: February 3rd! We have multiple tracks that should be of interest to both the ML community and experimentalists!
0
5
22
There’s really no better than feeling than when your first mentees get the @NSF GRFP! @garykbrixi, you’re going to continue to be a scientific rockstar in grad school! 🌟 You and @SabrinaKoseki are the most deserving recipients! I’m so proud of you guys. 💙.
1
0
22
Hi all, I'll be presenting our team's work on designing target-binding peptides with generative language models (and other cool protein design work!) tomorrow @valence_ai! 💻🧫 Please find Zoom details here: Looking forward to seeing you there! 🥳.
1
2
20
Beautiful beautiful work! I’ve said that pLDDT really tells you about how disordered proteins are — now we can leverage AF2 with simulations to explore it! 🥰 Would be excited to see this extend to AF2-Multimer as well. 😅.
Happy to share the publication of our paper in @Nature. Conformational ensembles of the human intrinsically disordered proteome. Work led by @GiulioTesei and @AnnaIdaTrolle. I'll post more later, but for now here is a link:.and a short movie about the work
0
1
20
Really excited to try this out! Btw, we should make it a norm for protein language models to be hosted on HuggingFace! 🤗.
🦅 the FAbCon has landed 🦅. Our #generative #antibody #LLM available on 🐣 FAbCon-small (144M).🐥 FAbCon-medium (297M) .🦅 FAbCon-large (2.4B). Team effort @alchemabtx @all_your_bayes @aretasgasp @jhrf @jakegalson @JorgeNDias 🎉.
1
3
20
Very excited to have our tour-de-force genetic screening paper out in @CellRepMethods! We've developed a powerful, integrated pipeline for identifying, screening, and interrogating genetic perturbations for defined phenotypic outputs. Let's review how STAMPScreen works! [1/5].
When scientists at the Wyss, @harvardmed, & @medialab got frustrated w/ existing tools & methods for gene engineering, they made their own. @PranamMIT, @krammecc, & colleagues invented STAMPScreen to help scientists get from a database to results fast.
3
5
18
@andrewwhite01 Miniproteins are this weird intermediate between the specificity of an antibody and the permeability/drug-like properties of peptides. I personally think miniproteins are a thing because diffusion/flow-matching are not ideal for antibodies or peptides. At least with the de novo.
0
1
17
An incredible paper that I have been excitedly anticipating is out in @Nature today -- so many congratulations to my brilliant friend and collaborator @MartinPacesa and the @MartinJinek lab on this groundbreaking structural elucidation of Cas9 conformational activation! 🎉🎊🎈.
Very excited to see our work on Cas9 conformational activation out in @Nature ! What started as a very poor crystal structure back in 2017 in the @MartinJinek lab, turned into a full blown cryoEM project during the pandemic.
0
0
14
@Pandeylab @NatureComms Do you understand how dangerous it is to draw strong conclusions from poorly analyzed data with severe methodological flaws? People look at journals such as @NatureComms for legitimate conclusions that can affect hiring, public practice, etc. This paper should be retracted!.
1
0
14
Hi all! I hope you can take a few minute to learn about our latest research on designing target-binding peptides (amongst other useful proteins) using #GenerativeAI! Thanks so much to @valence_ai for having me! 🙌🏾
0
2
15
Read more about what my company @GametoGen is up to: It's crazy to see how far it's come since @martinvars and I founded it back in 2020! 🤯 @DinaRadenkovic and @krammecc continue to lead it to newer heights -- you guys are amazing! 🌟.
1
0
14
Check out our new COVID-19 diagnostic platform, out today in @ScienceAdvances! Peptide beacons + mini-TIRF technology = sensitive S-RBD protein detection at femtomolar resolution. Really a great extension of our ubiquibody design work! 🤩
2
2
14
@martinvars @DinaRadenkovic @GametoGen An wonderful story by @Forbes of @DinaRadenkovic's vision and leadership for @GametoGen that led us here: 🎉.
0
1
13
Super excited about trying out CellREADR from our friends in Josh Huang's lab here at @DukeNeuro! This will be very useful for dynamic cell state engineering applications. 🧫
0
0
12
Hi everyone! We are hosting Duke AI Day (sponsored by.@nvidia) on our beautiful @DukeEngineering campus on June 7th!☀️If you work on developing or applying AI algorithms, we'd love for you to register (it's free to everyone!) and submit an abstract:
2
6
13
We are so grateful to #EndAxD for funding our research leveraging generative language models to design peptide-guided degraders of dysregulated GFAP! 🙏Continued research support will allow us to develop this into a potent and specific therapeutic for Alexander disease! Please
0
1
12
So excited to have @sacdallago and @AlexanderTong7 join us @DukeU!! 💙 We're building such an amazing AIxBio community with @rohitsingh8080 @romerolab1 .and others! 🙌 ESPECIALLY in all things protein/bio language models! 💻 🧬 Come join us in Durham! 😈.
Two major life updates: .- moving to Senior Applied Research Scientist in Digital Biology at NVIDIA (Jan '25).- starting a new lab at Duke as Visiting Prof (early '25) . Both roles focus on tackling hard problems in biological machine learning through collaborative research.
1
1
12
Check out our early-stage work on designing computationally-optimized peptides that bind to SARS-CoV-2 and recruit E3 Ubiquitin Ligases for degradation! Could be a potential alternative to PAC-MAN? #COVID19.
2
2
11
Okay, I know #AutoGPT is the AI hype right now, but check out the Regression Transformer! Discontinuous-masking-based autoregression for conditional protein sequence generation?! Yeah, my lab's definitely using this! 😎
1
2
12
Absolutely beautiful work from my talented and amazing friend, @JulesGrunewald!! If you're crazy about base editing, what's better than doing both C->T and A->G at the same time?! 🧬🍾.
2
1
9