Martin Pacesa
@MartinPacesa
Followers
3,833
Following
985
Media
314
Statuses
1,860
Structural biologist working on 🖥️ protein design, AI, and #CRISPR -Cas gene editing ✂️🧬 Avid weirdness connoisseur 🎩
Lausanne, Switzerland
Joined December 2018
Don't wanna be here?
Send us removal request.
Explore trending content on Musk Viewer
BLEACH
• 167020 Tweets
Saka
• 126035 Tweets
#SixTONESANN
• 73559 Tweets
#らんまアニメ
• 58259 Tweets
DARLING DARLING ITS HANNI DAY
• 50863 Tweets
Southampton
• 46461 Tweets
Havertz
• 34137 Tweets
Fulham
• 29811 Tweets
Sterling
• 23303 Tweets
ジェシー
• 22001 Tweets
Brentford
• 19345 Tweets
Martinelli
• 16502 Tweets
レインボーブリッジ封鎖
• 12805 Tweets
#オールスター後夜祭
• 11625 Tweets
Ahtisa
• 11147 Tweets
Kovacic
• 10575 Tweets
Last Seen Profiles
Pinned Tweet
Have you ever wanted to design protein binders with ease? Today we present 𝑩𝒊𝒏𝒅𝑪𝒓𝒂𝒇𝒕, a user-friendly and open-source pipeline that allows to anyone to create protein binders de novo with high experimental success rates.
@befcorreia
@sokrypton
24
399
2K
Today in
@Nature
we present the results of our exploration of the soluble protein universe and the design of functional soluble membrane proteins with
@CasperGoverde
@GoldbachNico
@befcorreia
1/6
21
217
915
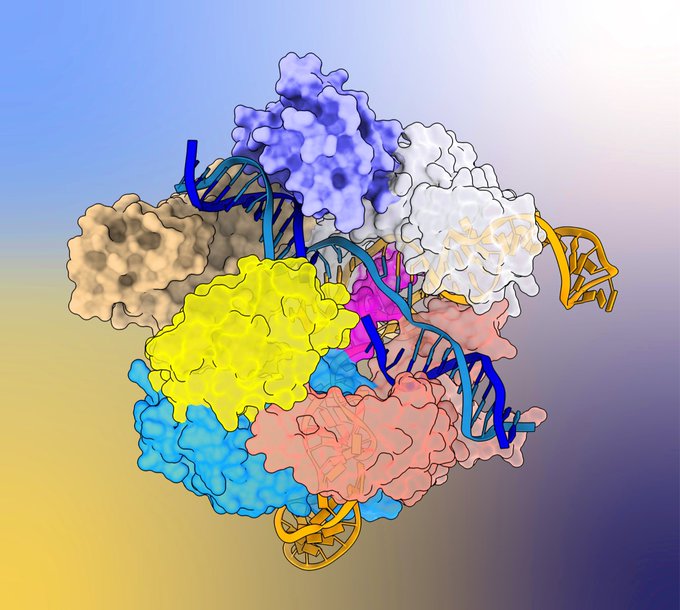
Very excited to see our work on Cas9 conformational activation out in
@Nature
!
What started as a very poor crystal structure back in 2017 in the
@MartinJinek
lab, turned into a full blown cryoEM project during the pandemic.
30
72
530
THEY SAID WE COULDN'T DO IT........well, no one actually said that. But in our updated manuscript with
@CasperGoverde
,
@GoldbachNico
, and
@befcorreia
we show you can now design soluble analogues of membrane proteins with preserved functional features!
9
92
490
I am happy to conclude my trilogy on Cas9 specificity with our new preprint from the
@MartinJinek
lab! We solved a staggering number (15!) of crystal structures of Cas9 bound to bona fide off-targets to investigate the nature of mismatch tolerance.
18
83
397
Although it is a very exciting development, it is sad to see that no code will be released, which would not be allowed for academic institutions
@nature
. It was clear that weights will not be released (despite being trained purely on public data) but this I did not expect.
Super excited to be releasing AlphaFold 3 today, developed by
@IsomorphicLabs
and
@GoogleDeepMind
: our next generation AI model for predicting the biomolecular structures and interactions of proteins, DNA, RNA, small molecules, and more:
1/
23
268
1K
4
47
241
This shit is both fucking hilarious and pretty annoying
2
23
237
Great work from
@ProfluentBio
on developing generative language models capable of structure informed enzyme and binder design! They leverage multi-state design to engineer functional variants of Cas9 and base editors.
2
48
230
Absolutely captivating article from
@fraser_lab
and Mark Murcko! Very down to earth discussion on what protein "structures" really are
2
51
222
My first publication from the
@MartinJinek
lab, in collaboration with
@CaribouBio
, is out now in
@MolecularCell
!
11
39
212
CRISPR gene editing has the potential to solve many problems in research, agriculture, and medicine. For the special 50th anniversary issue of
@CellCellPress
, we discuss its past, present and future directions with
@MartinJinek
and
@OanaPelea
.
3
61
204
Very happy to share the new manuscript from
@befcorreia
lab! With
@CasperGoverde
we de novo designed several complex natural folds, including soluble analogues of integral membrane proteins!
6
38
180
Wow, this is really unexpected! Designs were done together with Lennart Nickel in
@befcorreia
lab. Our pipeline will be fully open source and preprint online hopefully next week 🙂
Congrats to
@MartinPacesa
!!! The first two hits were pretty much identical to known binders so are not counted in the leaderboard
1
3
26
9
20
162
I am super excited to announce that I will be joining the lab of
@befcorreia
at
@EPFLEngineering
in Lausanne 🇨🇭 starting next month 🧬🔧 can’t wait to finally be part of the insanely thrilling field of protein design 💜
14
5
158
Keeping the ball rolling with my next manuscript from the
@MartinJinek
lab! We solved several
structures of intermediary binding states of Cas9 to elucidate the mechanism of target DNA binding and conformational activation!
12
30
136
We have updated our preprint with
@CasperGoverde
and
@befcorreia
on the design of soluble membrane protein analogues! We have now included proof of principle designs to show that our analogues can be functionalized via sequence grafting.
2
30
135
@Jiankui_He
Absolutely criminal, your lack of ethics and scientific rigour should preclude you from ever doing science again
5
5
128
Very glad I had the opportunity to work with a team from
@NobelPrize
and provide the structural renderings for the fantastic Chemistry prize poster!
2
15
124
After 5 years I have successfully defended my PhD at the
@UZH_en
in
@MartinJinek
’s lab! A big thanks to Martin, to everyone in the lab and those who attended the defence 🥳 it was a lot of fun and I hope it will continue to be in the future 🧬
21
1
118
New amazing research from the
@befcorreia
lab!
@zanderharteveld
and Alexandra explore protein topologies that have not been found in nature using a novel deep learning pipeline called Genesis
2
33
117
Your daily reminder that packaged virions sustain internal pressures of up to 10 bar, which is 5-times more than your car tire.
Wanted to again highlight these beautiful simulations by
@aksimentievLab
2
21
109
Revolutionary work from the
@ProfluentBio
team 💻🔥🧬 not only is OpenCRISPR fully AI-generated and free to use; it also matches the efficiency of SpCas9, has much better specificity, and is also compatible with all Cas9-based technologies like base editing ✂️ amazing!
Can AI rewrite our human genome? ⌨️🧬
Today, we announce the successful editing of DNA in human cells with gene editors fully designed with AI. Not only that, we've decided to freely release the molecules under the
@ProfluentBio
OpenCRISPR initiative.
Lots to unpack👇
281
979
4K
2
14
108
Very glad to see this paper from the
@palermo_lab
out, congratulations! These guys are absolute wizards when it comes to MD! Big shout out to
@pablitoarantes
@aakashsahha
@MartinJinek
and my student Jonas Binz!!
5
14
96
We have updated our bioRxiv preprint on Cas9 off-target activity! What is new you may ask? First of all, much sleeker and better looking figures and readability that make this colossus more accessible!
I am happy to conclude my trilogy on Cas9 specificity with our new preprint from the
@MartinJinek
lab! We solved a staggering number (15!) of crystal structures of Cas9 bound to bona fide off-targets to investigate the nature of mismatch tolerance.
18
83
397
4
14
92
I have made a user-friendly Colab notebook that allows generation of novel protein sequences based on the fantastic ProtGPT2 model from
@ferruz_noelia
. Give it a go and let me know if there are any problems!
1
10
87
Fantastic new preprint out from the
@befcorreia
lab, demonstrating we can invert the AlphaFold network to design de novo proteins!
@CasperGoverde
showing off his AF2 hacking skills 😁
De novo protein design by inversion of the AlphaFold structure prediction network
#biorxiv_bioinfo
0
14
51
1
8
76
New surface-based PPI design pipeline from the
@befcorreia
lab!
Our paper with
@befcorreia
@CirauquiPablo
et al on protein design using geometric deep learning is finally in
@nature
We show experimental results of diverse de novo designed binders
28
160
712
0
8
60
Another
#ModelAngelo
success! It managed to build 80% of the very loopy structural model into the 3.3A Glacios 200 keV map. Although may not look impressive at first, it allowed me to align individual AlphaFold predicted domains and fit perfectly. Full model done in 20 minutes
2
8
60
Hey
#Science
Twitter, I just had my whole genome sequenced, what are some cool things to try/check out? 🧬🧬
10
1
54
I gave this ago and I am pleasently surprised by the results! It is able to accurately predict the pseudo knot structure of the Cas12b sgRNA! Only the first stem loop is predicted to be unstructured even though it forms a hairpin. Purple is prediction, salmon ground truth
De Novo RNA Tertiary Structure Prediction at Atomic Resolution Using Geometric Potentials from Deep Learning
#biorxiv_bioinfo
1
9
23
1
4
54
Super important work from the Levy lab! Predicted oligomeric protein assemblies from model organisms to overcome the huge problem of the AlphaFold database, where all proteins are predicted as monomers, which is not the case most of the time in vivo!
⚛️Protein structures are key to elucidating molecular details of cellular processes.
#AlphaFold2
& others unveiled millions of tertiary structures, but these proteins' quaternary structures remain mostly a mystery. Here's how to bridge the gap. Thread👇
4
120
396
1
8
51
Stunning performance! Modelled entire Cas9 correctly in 13 minutes! Tested on 3.0A map of EMD 14499, 7Z4J PDB for comparison in grey.
We are proud to present
#ModelAngelo
: a deep-learning based approach to automated model building in
#cryoEM
maps by the amazing Kiarash Jamali (
@jamaliki1998
), with help from Dari
@kimanius
.
17
174
705
1
6
47
I am so thrilled to have had the opportunity to give a seminar about my PhD work on Cas9 specificity at
@AstraZeneca
. The discussions and feedback were amazing! Looking forward to seeing more transformative work from their labs!
1
2
47
@fermatslibrary
Oh gosh, just think how much time I could have saved if I knew this handy trick before!
0
0
46
So happy to see this monumental work from
@ElevyLab
out in
@CellCellPress
today! You can use the database to see if your protein of interest is likely to oligomerise and then use it to rationalise pathogenic mutations, which often occur at oligomer interfaces.
So happy to see our work published at
@CellCellPress
!
We added the quaternary structure dimension to proteome-wide,
#AlphaFold2
predictions:
The published paper:
The original 🧵 (Two more experimental structures since!)
7
109
307
3
9
46
@MoAlQuraishi
Not releasing weights was expected but not releasing even code is pretty bad. What is the point of publishing in a scientific journal then, as opposed to the existing white paper? This is pretty close to a paid promotion imho
2
1
45
Lastly, I would like to share the cover design I made for this article, that sadly did not make it into print. It shows a children’s shape matching toy, where we force the wrong shapes into the holes, similar to what Cas9 does to mismatched base pairs! Hand of
@m_cherepkova
😁
2
2
40
What this guy does with deep learning and proteins is magic, well deserved position Sergey!
I'm excited to share that I'll be joining
@MITBiology
as an Asst Prof. in Jan 2024! Come join us! 🤓🧪🖥️🧬
178
156
2K
1
1
43
@ddoniolvalcroze
Morrissey is for me the perfect example when explaining “Love the art, hate the artist”
0
3
39
Absolutely incredible structures, right from its native membrane! Stunning work
1
2
39
We tried to solubilize the claudin, rhomboid, and GPCR membrane folds. Using vanilla ProteinMPNN, we were not able to design soluble versions of these proteins, so
@JustasDauparas
trained a new version called SolubleMPNN resulted in many soluble designs of these folds.
1
5
39
I finally had some time to put up these wonderful prints of molecular art by
@fenguita
. I am particularly proud of the two renderings of my own Cas9 structures, for which I am very thankful to him! Go check out his amazing art and support him at
!
1
7
39
Why don’t we get paid for each citation and download of our paper, kinda like musicians make money on Spotify
#StreamingGoogleScholar
7
4
37
Go check out Kim’s great podcast, where I got to rant about my past and present research, and the outstanding problems in CRISPR and protein design! ✂️🖥️🧬
Got weekend plans? You do now! Check out the latest episode of
#CRISPRli
. Join our conversation as
@MartinPacesa
from
@EPFL_en
and I enter the world of Cas9 structures and computational protein design. Don't miss out... 🎙️
#CRISPR
#proteindesign
0
9
33
0
3
35
Very excited about these phase 1 results from
@CaribouBio
! The ultra high specificity of the chRDNA platform was crucial for achieving this. Very glad we had the opportunity to collaborate and elucidate the mechanism of increased specificity.
0
2
34
Come check out my talk on Monday
@harvardmed
on the mechanism of Cas9 activity!
1
5
33
The
@befcorreia
lab is looking for a postdoc! As a postdoc there myself, I can tell you that Lausanne is a stunning city, EPFL has a strong collaborative environment, and the lab researchis super exciting! On top of that, Bruno is a really chill dude 😎
1
12
32
Btw, if anyone can figure out what the protein on the right does (Uniprot Q8U0N8) then let me know! It has been haunting me for months
8
4
32
This is an absolutely stunning animation 😍😍 now I wanna see an adenovirus in there
Receptor/Clathrin-mediated endocytosis. The challenge was to get the flat membrane 🟦 to curl into a vesicle, but it worked using
#geometrynodes
! TfR 🟧, Clathrin 🟩. For the clathrin simulation, I used
@bradyajohnston
's add-on.
#SciComm
#SciArt
#b3d
#molecularanimation
25
461
3K
0
4
31
@jankosinski
Wasn’t AF3 trained in the Jasper database? Maybe the sequence was part of the training
1
2
30
Really nice read! Investigating the collateral trans cleavage activity of Cas12a in mouse embryos. Can some of the more experienced in vivo gene editing folks comment on how robust the method is?
#CRISPR
#GeneEditing
1
3
30
What an amazing event
#ERCL23
was! Lots of exciting research, familiar faces, and great evening programme 🎉 4 talks from the
@befcorreia
lab!!! Thanks to everyone who stopped by for the discussion and super nice feedback on our project with
@CasperGoverde
!
0
2
30
Such a cool effort to make cryoEM more high throughput!
EasyGrid: A versatile platform for automated cryo-EM sample preparation and quality control
#biorxiv_molbio
0
14
44
1
2
28
He finally emerges from the shadows 🎉
First tweet ever. Big congratulations to Jennifer Doudna
@doudna_lab
and Emmanuelle Charpentier
@manue_lab
on their Nobel Prize!
31
52
912
2
0
26