Valentina Giunchiglia
@valegiunca
Followers
203
Following
560
Statuses
153
PhD student @ImperialMed and visiting researcher @HarvardDBMI, Artificial Intelligence and Computational Neuroscience
Joined May 2021
⭐️LOGML is back and we are looking for mentors!⭐️ Do you work in geometry or machine learning? Apply by February 16th! Mentors lead a group of 6 students to work on a project during the week and some groups end up publishing their work. More information at
⭐️Mentor applications open⭐️ We're excited to announce that LOGML summer school will return in London: July 7-11 2025. We are seeking mentors to lead group projects at the intersection of geometry and machine learning. Find out more and apply:
0
3
13
RT @LogmlSchool: Do you work in geometry and / or machine learning? Do you want to help us make @LogmlSchool an unforgettable experience?…
0
9
0
RT @marinkazitnik: Excited about the bright future of KG + LLM-based AI agents! Congrats to @xiaorui_su, @GaoShanghua, and @valegiunca on a…
0
11
0
RT @LogmlSchool: ⭐️Mentor applications open⭐️ We're excited to announce that LOGML summer school will return in London: July 7-11 2025. W…
0
30
0
RT @KempnerInst: NEW in the #KempnerInstitute blog: learn about ProCyon, a multimodal foundation model to model, generate & predict protei…
0
11
0
This was a lot of work! ProCyon is a model that integrates protein sequences, structures, and natural language to predict and generate protein phenotypes Check the paper @ImperialMed @KingsCollegeLon @imperial_mrcdtp @KingsIoPPN @imperialcollege
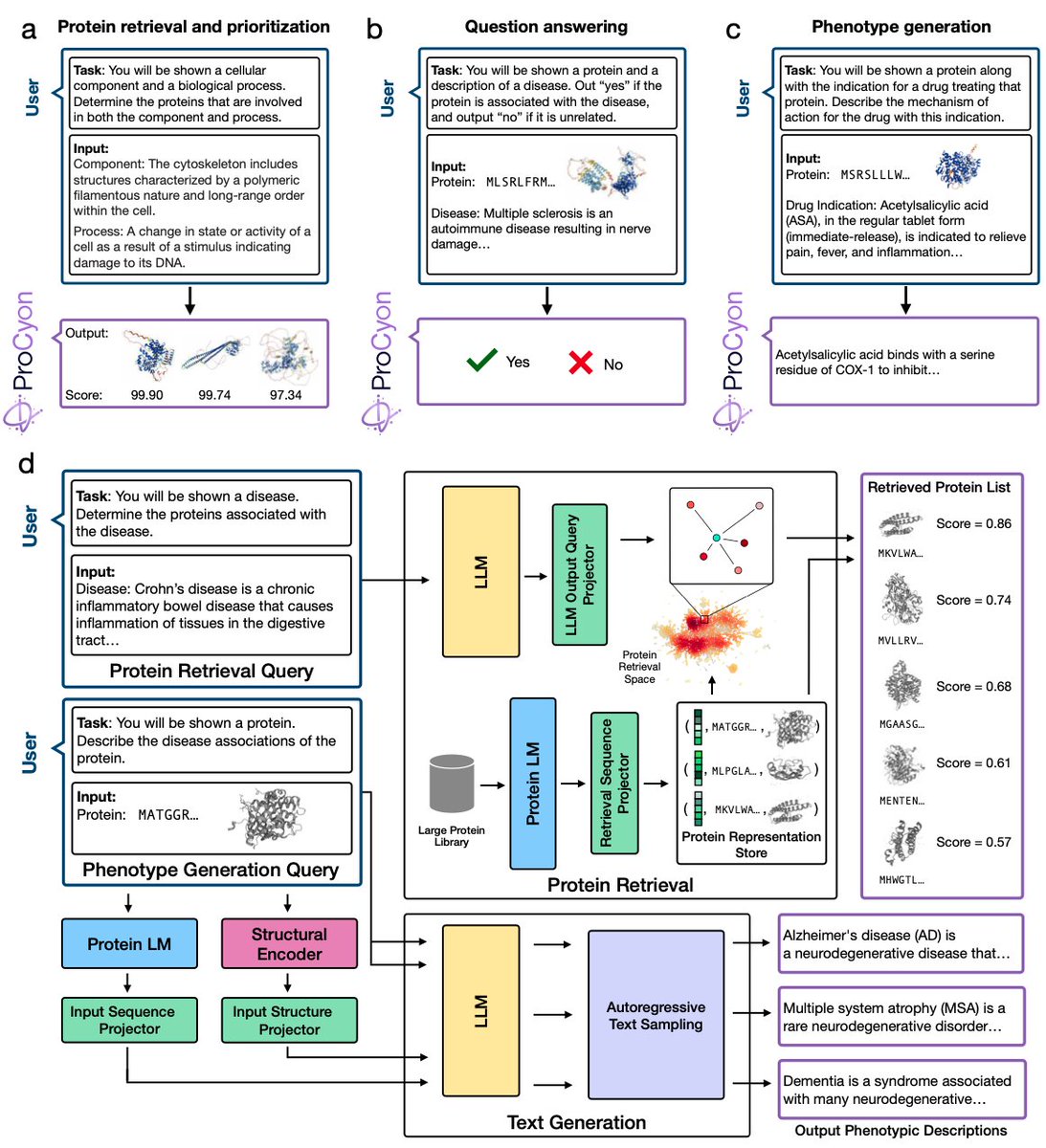
(1/4) Excited to introduce ProCyon: a multimodal foundation model to model, generate, and predict protein phenotypes, led by stellar @oq_35 @YepHuang Robert Calef @valegiunca 👉 ProCyon is an 11B parameter multimodal model that integrates protein sequences, structures, and natural language ProCyon models, predicts, and generates phenotypes with zero-shot task transfer, breaking past the limits of fixed vocabularies or ontologies 🔬 The Challenge: Predicting protein structure is advanced, but predicting protein phenotypes—observable traits linking molecular function to biology—is still an open problem. 40% of human proteins lack context-specific insights, while 20% remain almost entirely uncharacterized
0
2
10
RT @marinkazitnik: (1/4) Excited to introduce ProCyon: a multimodal foundation model to model, generate, and predict protein phenotypes, le…
0
28
0
RT @BiologyAIDaily: PROCYON: A multimodal foundation model for protein phenotypes • Introducing PROCYON, a groundbreaking multimodal model…
0
4
0
RT @marinkazitnik: Medicine thrives on knowledge, yet clinical vocabularies are fragmented. AI struggles to unify this knowledge, creating…
0
64
0
RT @marinkazitnik: Are biomedical AI models truly as smart as they seem? @YEktefaie Our @NatMachIntell paper intr…
0
37
0
RT @npjDigitalMed: New study introduces IDoCT, a breakthrough method enhancing the precision of online cognitive tasks by addressing device…
0
1
0
RT @ProfHampshire: Great to see this innovative work by @valegiunca published in npj Digital Medicine! She developed a flexible new approac…
0
1
0
Thanks to all my collaborators @DragosCGruia @LeredeAnnalaura @neurohellyer William Trender, and @ProfHampshire for the supervision! @imperial_mrcdtp @KingsCollegeLon @ImperialMed @KingsIoPPN @iamcognitron
3
0
0
Check out our paper on KGARevion, a KG+LLM agent 🤖designed for knowledge-intensive medical QA A summary of the paper is available here @imperial_mrcdtp @ImperialMed
Introducing KGARevion, a KG+LLM agent 🤖designed for knowledge-intensive medical QA. Led by @xiaorui_su @GaoShanghua @valegiunca Here's why it is unique: 1️⃣ Open-ended reasoning: Generates nuanced answers without relying on predefined options 2️⃣ Robustness: LLMs can be surprisingly sensitive to how candidate answers are ordered and indexed in multi-choice setups Unlike LLMs, which show order sensitivity in multi-choice setups (favoring answers in the first position!), KGARevion is immune to this bias, ensuring more reliable selections 3️⃣ Flexibility: Combines LLM-generated triplets with KGs for verified answers. The multi-step process makes KGARevion adaptable to different reasoning styles, outperforming RAG models Medicine relies on various reasoning strategies, including rule-based, prototype-based, case-based, and analogy-based vertical reasoning (Patel et al., 2005). This variety calls for approaches that accommodate different styles and integrate specialized, in-domain knowledge. For instance, an organism such as Drosophila is used as an exemplar to model a disease mechanism, which is then applied by analogy to other organisms, including humans. In clinical practice, the patient serves as an exemplar, with generalizations drawn from many overlapping disease models and similar patient populations. 4️⃣ Versatility: Works with different LLMs and medical KGs. Preprint: Thanks Discover AI for highlighting KGARevion: @HarvardDBMI @harvard_data @KempnerInst
0
1
8
Our recent perspective has been published in @CellCellPress! Check out our paper where we discuss "AI scientists" as collaborative AI agents to empower biomedical research. @imperial_mrcdtp @ImperialMed
Excited to share our perspective in @CellCellPress, where we discuss “AI scientists” as collaborative AI agents designed to empower biomedical research While the concept of an “AI scientist” is aspirational, advances in agent-based AI are paving the way for AI agents as conversable systems with reflective and reasoning abilities that coordinate LLMs, ML tools, experimental platforms, or combinations thereof. We outline initial autonomy levels for agents based on proficiency in hypothesis generation, experimental design, execution, and reasoning: Level 0: No AI agents Level 1: AI agents as research assistants Level 2: AI agents as collaborators Level 3: AI agents as scientists These “AI scientists” could enhance discovery workflows by introducing skepticism and reasoning. Our vision is to amplify human creativity, enabling AI to handle large datasets, navigate complex hypotheses, and perform repetitive tasks faster. Many ethical considerations arise with biomedical AI agents. We discuss issues of governance, robustness, evaluation protocols, dataset generation, and associated risks. Imagine AI agents that help: 🔬 Design discovery workflows 🌱 Simulate virtual cells 🎛️ Control phenotypes programmatically ⚙️ Design cellular circuits 💊 Optimize therapeutic discovery and development Many thanks to fantastic group of co-authors @GaoShanghua, @AdaFang_ @YepHuang, @valegiunca, @ayushnoori, @schwarzjn_, @YEktefaie, @kondic_jovana
@HarvardDBMI @harvardmed @KempnerInst @harvard_data @broadinstitute
0
0
11