Sukrit Singh
@sukritsingh92
Followers
1K
Following
55K
Statuses
3K
Biophysicist|@DamonRunyon QB + @theNCI K99/R00 Fellow @MSKCancerCenter w @jchodera | @foldingathome|PhD w @drGregBowman|resistance, dynamics, dogs 🇮🇳🇸🇬🇺🇸
nyc
Joined June 2016
RT @XuhuiHuangChem: Exciting news! Our TS-DAR method is now published in @NatureComms! 🎉 TS-DAR identifies transition states across multipl…
0
18
0
RT @foldingathome: Too many requests for money this Giving Tuesday? Consider giving your spare CPU/GPU cycles to advance biomedical resear…
0
10
0
Shoutout to citizen-scientists @foldingathome where we ran work units to retrospectively analyze the "really bad" predictions, and identify ways we can improve them!
0
1
3
RT @JoshRackers: We can accelerate molecular conformational ensemble sampling with generative AI! Walk-Jump Sampling smoothes out the dist…
0
13
0
RT @mia_rosenfeld: wow, what an amazing and massively useful guide to large parallel simulations from @sukritsingh92 and @sonyahans !!!! 🤩💻
0
1
0
Excited to share my and @sonyahans's little guide to large parallel simulations (and what you can do with them) in preprint form!
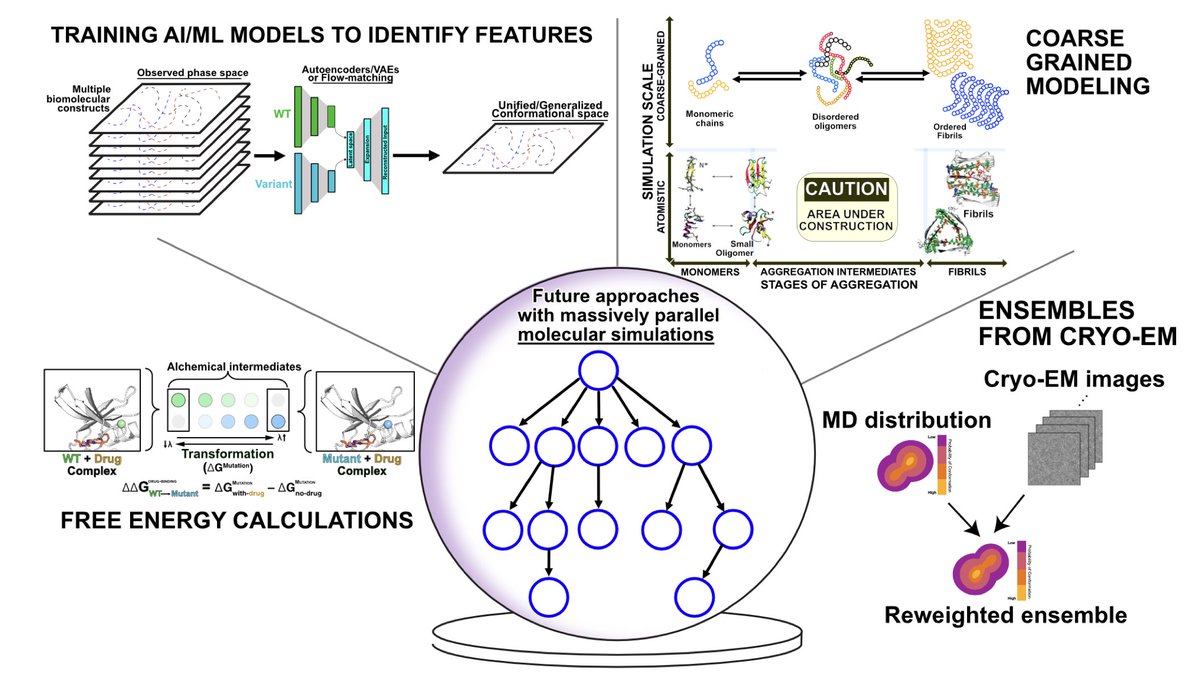
Running and Analyzing Massively Parallel Molecular Simulations 1. This article provides a comprehensive guide on generating and analyzing massively parallel molecular dynamics (MD) simulations to explore protein conformational landscapes and their role in biological processes. 2. Massively parallel simulations overcome the limitations of single MD simulations by sampling long-timescale events and complex biological phenomena through distributed computing. 3. The paper highlights the use of platforms like Folding@home to create datasets from thousands of citizen-scientist-powered simulations, capturing entire energy landscapes of proteins at unprecedented scales. 4. It emphasizes key practical considerations, including selecting appropriate force fields, handling storage needs, and configuring simulations to maintain consistency across datasets for future research comparisons. 5. The guide introduces advanced methods such as adaptive sampling and replica exchange, which improve the exploration of rare events and enhance predictive power for therapeutic design. 6. The analysis section details how Markov state models (MSMs) and time-lagged independent component analysis (tICA) enable insights into protein dynamics, allostery, and mutational impacts relevant to health and disease. 7. Tools like OpenMM and GROMACS are recommended for running large-scale MD simulations, with detailed advice on handling complex systems such as membrane proteins or protein-ligand complexes. 8. The article also explores novel strategies for handling computational challenges, such as varying initial configurations and using adaptive seeding to ensure robust sampling. 💻Code: 📜Paper:
0
13
34
RT @BiologyAIDaily: Running and Analyzing Massively Parallel Molecular Simulations 1. This article provides a comprehensive guide on gener…
0
14
0
RT @sonyahans: The Conference for Undergraduate Women and Gender Minorities in Physics (CU*iP) application deadline is Oct 23rd! NYU is one…
0
4
0
RT @MarkHahnel: Big shout out to those researchers who made their data available in PDB over the years The Chemistry Nobel Prize winners t…
0
225
0
RT @RolandDunbrack: Why Alphafold succeeded - the best large, curated, cleaned training data set is the PDB. Thank Helen Berman for that.
0
25
0
RT @ivyzhang__: Registration is now open for the next CADD GRS, taking place at the University of Southern Maine July 12-13, 2025! Great…
0
7
0
RT @MSKEducation: We want to thank @MSKCancerCenter Communication for helping us bring the stories behind our scientists and giving us an i…
0
4
0
RT @gonenlab: Folks please remember, when looking for a lab to join, always talk with current *and* past trainees. Joining a lab that nurt…
0
12
0
RT @tiwarylab: Presenting our frank perspective on some impressive achievements of Generative AI for CompChem & how far it remains to go ht…
0
27
0
@drGregBowman Might be different saving frequencies which would change the storage footprint, no? Could be even more!
1
0
1