Jack Bravo
@jpkbravo
Followers
1,033
Following
736
Media
41
Statuses
405
Assistant Professor at IST Austria | Studying nucleoprotein complexes one particle at a time
Joined April 2018
Don't wanna be here?
Send us removal request.
Explore trending content on Musk Viewer
Colombia
• 365334 Tweets
Uruguay
• 272175 Tweets
Seleção
• 270937 Tweets
七夕の日
• 193993 Tweets
都知事選の投票
• 172454 Tweets
Neymar
• 142540 Tweets
Brazil
• 129203 Tweets
織姫と彦星
• 114849 Tweets
Bielsa
• 82499 Tweets
Tank
• 77662 Tweets
Endrick
• 66727 Tweets
#七夕の願い事
• 64628 Tweets
Militão
• 64271 Tweets
Dorival
• 62714 Tweets
Rafael
• 60151 Tweets
Shakur
• 54029 Tweets
भगवान जगन्नाथ
• 52475 Tweets
Alisson
• 51296 Tweets
BELLElieve ON ASAP
• 34615 Tweets
Jai Jagannath
• 33826 Tweets
Rodrygo
• 29821 Tweets
Rochet
• 29616 Tweets
静岡40度
• 28825 Tweets
プロキオンS
• 28467 Tweets
#DiazMasvidal
• 27995 Tweets
#ABCお笑いグランプリ
• 25425 Tweets
Ronaldinho
• 22382 Tweets
#短冊メーカー
• 19777 Tweets
Douglas Luiz
• 18994 Tweets
Danilo
• 18465 Tweets
願いごと
• 16981 Tweets
Arrascaeta
• 13595 Tweets
ヤマニンウルス
• 12887 Tweets
Last Seen Profiles
Pinned Tweet
I'm very excited to announce that I'll be starting my lab at
@ISTAustria
near Vienna this summer! The Bravo lab will be using enzyme kinetics and cryo-EM to study bacterial immunity. Read more here:
46
36
336
1. I am thrilled to share the accepted version of our Cas9 mismatch manuscript, now in
@Nature
! Here’s a (not-so) brief outline of some of the findings from our original preprint, and some VERY exciting new findings
16
119
499
I am thrilled to share my latest work, out today in
@Nature
. This project started ~18 months ago as a blueprint for my own lab, which will start at
@ISTAustria
in July, but not have been possible without the support of
@davidwtaylorjr
& the whole team
13
42
248
1. I am delighted to be able to share our story on the enigmatic DISARM antiphage defence system, in collaboration with
@davidwtaylorjr
,
@_aparicioC
,
@NobregaFL
and
@StanBrouns
5
29
101
More exciting news - our type III-B CRISPR protease signalling story is published! Congrats to my co-first authors Jurre and
@crp_salazar
, the rest of the team led by Raymond Staals
Another cool story to share before
@crispr2023
- this time in collaboration with Jurre Steens, Raymond Staals & others: how cOA binding activates a CRISPR-associated protease to activate an immune signalling pathway
3
17
78
3
19
81
Another cool story to share before
@crispr2023
- this time in collaboration with Jurre Steens, Raymond Staals & others: how cOA binding activates a CRISPR-associated protease to activate an immune signalling pathway
3
17
78
It's great to be able to share the final version of this story, led by
@Jamie_Yelland
along with myself,
@DrJoshBlack
@ArlenJohnson8
&
@davidwtaylorjr
. We found a pretty neat mechanism of how a single ribosomal methylation regulates ribosomal maturation
2
13
68
I'm so happy to share our SpRY-Cas9 story in its final form! Congratulations to first author
@GraceHibshman
- it was a great collaboration with a diverse team led by
@davidwtaylorjr
, including
@IlyaFinkelstein
and many others... 1/n
9
13
66
Very pleased to share our work on the type II-D CRISPR effector!
Check out our new work by fabulous grad student Rodrigo
@TexasScience
@ut_mbs
! Guide RNA folds into gnarly geometry to tether tiny protein domains together. Fun collaboration with
@metagenomi
! DNA targeting by compact Cas9d and its resurrected ancestor
4
18
71
2
5
64
I’m thrilled to be able to share this really exciting story that I’ve been working on for the last couple of years with
@GraceHibshman
on how an engineered Cas9 variant can identify targets WITHOUT the need for a specific PAM sequence. 1/n
3
8
57
My group at
@ISTAustria
is recruiting a lab manager/research technician to join when we start in July! If you're passionate about science and thrive in a dynamic environment, apply here:
#ResearchJobs
#LabManagement
#ScienceCareers
0
24
35
14. Please read the paper for many more details on Cas9 mechanisms. I’d like to thank
@davidtaylorjr
for his support, and the tireless work of
@GraceHibshman
for finding the right SuperFi mutants. This was a great story to be part of, and there’s a lot more cool stuff to come
1
0
33
Great to see this out! Always a great time working with the dream team
@davidwtaylorjr
@PeterFineran
@palermo_lab
@RobFagerlund
and my co-first authors
@EvanASchwartz
and Mohd Ahsan.
Great work by former grad student
@EvanASchwartz
and postdoc
@jpkbravo
@TexasScience
@ut_mbs
and Mohd Ahsan
@palermo_lab
in
@NatureComms
! Couldn’t happen without long time collaborators
@RobFagerlund
and
@PeterFineran
@otago
.
0
12
44
3
4
33
Another day, another seriously odd CRISPR complex! Great story about a weird partially-fused type III effector complex. A weird evolutionary stepping stone between III-A and III-E perhaps? Congrats to
@EvanASchwartz
@RobFagerlund
@PeterFineran
et al.!
🚨Pre-Print Alert🚨 Happy to share our new story on the type III-Dv system from Synechocystis🦠-my favourite organism😍 Another great collaboration with
@davidwtaylorjr
! Also with
@PeterFineran
,
@jpkbravo
, Brodbelt Lab and structural wiz
@EvanASchwartz
!
2
9
41
0
4
28
This is a really superb piece of work, I’m so glad to see it published! Congrats
@PeterFineran
@rafapinilla92
@SarahCamara94
at el!
1
2
26
This is a really important, nuanced study from
@LarissaKruger93
&
@mfwhite2
- CBASS is inhibited through conjugation to a conserved, phage shock host protein, and the cyclase module is released upon phage infection by Cap3. What an elegant strategy for activation!
Reversible conjugation of a CBASS nucleotide cyclase regulates immune response to phage infection
#biorxiv_micrbio
0
8
37
0
3
23
Great to finally have this work out - we solved 6 structures of Cas9 at with different mismatches revealing why certain mismatches are tolerated while others block Cas9 activation!
2
3
22
So cool that we got the cover of the current issue of
@NatureSMB
! Congrats again to
@Jamie_Yelland
and the whole team
❄️our January issue is live❄️
on the cover we feature work from
@davidwtaylorjr
&
@ArlenJohnson8
labs on how a single 2′-O-methylation of rRNA gates assembly of a functional ribosome
3
7
45
0
1
21
What a week for anti-phage defence systems! The imminent Beyond CRISPR EMBO workshop should be a very exciting meeting
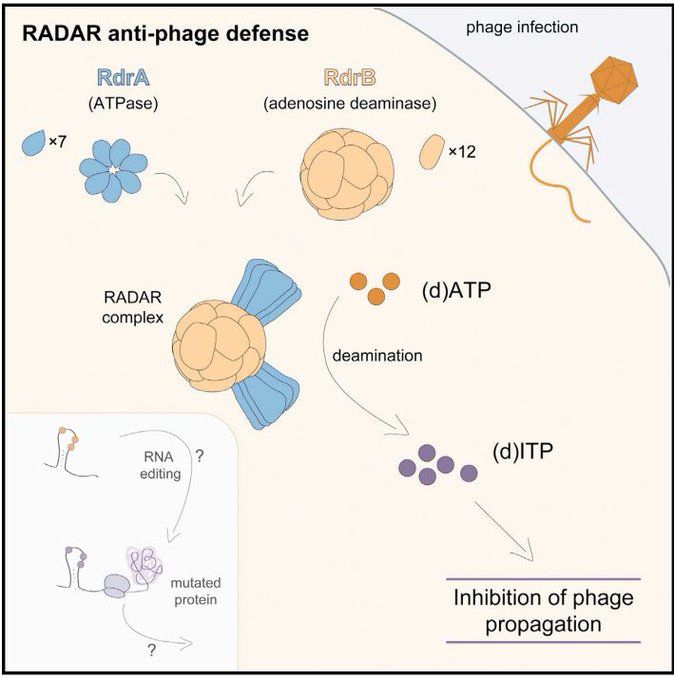
Out
@CellCellPress
: Together with the Kranzusch lab, we characterized RADAR, a giant immune machine that blocks viruses via adenine deamination
A complex of 96 protein subunits, 4 times heavier than the ribosome
Congrats
@B_DuncanLowey
and
@Nitzan_T
!
2
112
411
1
2
20
Great to have this story out. Congrats to my co-first authors
@EvanASchwartz
&
@Tess_M_McBride
and other collaborators!
Great work from grad students
@EvanASchwartz
@UTexasILS
@TexasScience
and
@Tess_M_McBride
@otago
about I-D Cascade picking targets
@NatureComms
. And another one - with
@PeterFineran
and
@RobFagerlund
.
1
9
63
1
4
18
I'm really looking forward to present some of my recent work at the
@TorontoRNA
meeting on Friday!
Start off your September with great talks from Dr. Jack Bravo
@jpkbravo
(Post-doc from the University of Texas-Austin) and Syed Nabeel-Shah (PhD candidate from University of Toronto) next Friday Sep 9 at 4pm EDT. Check out the poster for more details! Zoom links will be emailed!
1
4
14
0
3
18
Very nice work on an anti-anti-phage mechanism
0
3
17
@kingcada
@lisaeshunwilson
@lindsey__young
@kylelmorris
@FrommSimon
@dabiophysicist
@BJ_Greber
It’s PNPase! Here’s an image from Dendooven et al.
4
3
17
There are loads of rRNA modifications, but what is their actual function?
@Jamie_Yelland
found a surprising role for one such modification in ribosome assembly.
I'm proud to be part of this great story with
@ArlenJohnson8
@DrJoshBlack
&
@davidwtaylorjr
0
4
16
Very proud of this story. We used cryoEM and kinetics to demonstrate how remdesivir inhibits the SARS-CoV-2 RdRp. Despite different experimental strategies, our results are in good agreement with those from
@CramerLab
Structure and kinetics of an elongation intermediate of SARS CoV-2 RdRp stalled by remdesivir. Great collaboration between
@jpkbravo
in my lab with
@dangerzoneTD
and Ken Johnson
@TexasScience
!
1
5
27
2
0
15
Excited to share a new pre-print from my post-doctoral work in the
@MartinJinek
lab at
@UZH_Science
together with the lab of
@Mblokesch
at
@EPFL_en
.
We describe the molecular mechanism of plasmid elimination by DdmDE defense system.
A long 🧵
7
51
165
0
1
14
This is a great summary of our recent work!
Defending against plasmids
#ResearchHighlight
This study reports the mechanism of plasmid clearance by the DNA defence module DdmDE of the Vibrio cholerae seventh pandemic strain.
0
5
15
0
0
13
Great to see such exciting work on the Zorya defense system. What a neat mechanism - phage genome injection triggers recruitment of a nuclease to degrade DNA as it enters the bacteria.
Very excited to release our Zorya anti-phage defense system preprint
@HaidaiHu
et al. Zorya has a rotary molecular motor with a 700 Å (!) tail. Wonderful collaboration with
@SimonJacksonNZ
,
@Salmo_Lab
, Richard Berry,
@thenielsenlab
and
@yongwangKU
#cryoEM
8
40
160
0
0
13
I'm super proud that this is my first paper as a corresponding author, and I'm very grateful for the support of my mentor
@davidwtaylorjr
, and the hard work of
@GraceHibshman
and the rest of the team.
3
0
13
Really proud of this story! We found that our favourite RNA chaperone protein NSP2 employs a surprising autoregulatory mechanism to prevent itself getting tangled up in large RNA-RNA networks
1
0
12
It's great to be able to share this work, led by
@Roisin_OBrien_
- some big surprises in here, including how AcrIF2 can inhibit both type I-C and I-F Cascades despite their structural differences
Excellent work from my grad student, now postdoc
@Roisin_OBrien_
and key lab members
@jpkbravo
@GraceHibshman
@jacquelyn_wrong
all
@TexasScience
! Structural snapshots of R-loop formation by a type I-C CRISPR Cascade
0
4
26
1
1
10
Absolutely stoked to share this work - a neat combination of cryo-EM, single-molecule fluorescence, HDX, UVXL, misc biochemistry and in vivo assays used to provide mechanistic insights into what started as an unexpected result in a control experiment...
Super-proud to share our work on rotavirus RNA chaperone published in PNAS
@PNASNews
Beautiful work spanning cryo-EM, single-molecule biophysics, structural proteomics and virology from
@jpkbravo
@BartnikKira
@anton_calabrese
@DavidovichLab
and many other talented co-workers!
7
6
75
0
2
10
Really wonderful to see our work out in
@MolecularCell
!
Excited to share our story about how Remdesivir inhibits SARS CoV-2 viral replication in
@MolecularCell
. Great work by my postdoc
@jpkbravo
and grad student
@dangerzoneTD
with Ken Johnson
@TexasScience
!
2
14
91
2
0
10
I promise this is the last manuscript we share for a little while (at least a week or two)... I think this takes
@davidwtaylorjr
lab up to 10 new structures shared this week. A great way to wrap up a wonderful
#CRISPR2022
meeting!
0
0
10
Our work is accompanied with another manuscript focusing on functional characterisation of Cas12a2 by
@Okrety_MD
@BeiselLab
@Jackson_Lab_USU
@tjHallmark
1
0
9
Check it out! A great global collaboration with
@BartnikKira
,
@DavidovichLab
,
@anton_calabrese
led by
@AlexBoRNA
.
0
1
7
Very exciting work...it looks like some CBASS systems were dsRNA sensors all along?
Congrats to
@daltonbanh
and
@camgroberts
for their work that found how a CBASS cyclase binds phage RNA to initiate immunity. Big thanks to our collaborators Sean Brady and Adrian Morales Amador
2
45
131
0
0
7
Holy shit MC Hammer just retweeted our preprint
@AlexBoRNA
Structural basis of rotavirus RNA chaperone displacement and RNA annealing | bioRxiv
#Hamm400aos
0
3
19
1
0
8
Thanks to my co-authors
@davidwtaylorjr
@Jackson_Lab_USU
@tjHallmark
@BeiselLab
- this has been an incredibly exciting collaboration with lots of surprises along the way. It was great to be able to give a little preview yesterday at
#CRISPR22
yesterday.
4
1
8
By using an autoinhibitory TL to block DNA binding, DrmAB can only bind ssDNA or 5’-ovh DNA typically found in phage genomes prior to circularisation and at sites of rolling circle DNA replication.
@gaofir
previously showed that injection and replication are targets of DISARM.
1
0
7
@AlexisRohou
You’d think that NMR would be surviving a bit better given that it’s solution technique though...
1
0
7
In an accompanying manuscript, the
@BeiselLab
&
@Jackson_Lab_USU
demonstrate that Cas12a2 triggers abortive infection in response to target RNA recognition, which occurs through non-specific dsDNA destruction. /5
1
1
7
I'd finally like to thank
@davidwtaylorjr
for his support, and our collaborators
@tjHallmark
@Jackson_Lab_USU
@BeiselLab
- it's been a real pleasure working on this project! It took >1yr to find suitable freezing conditions, and each structure was more surprising than the last.
0
0
6
@BKleinstiver
One particularly cool aspect of this story was that we used kinetics analysis to guide cryo-EM sample prep to capture R-loop intermediates. A single dataset yielded 7 structures (2.8 - 3.3Å)! 4/n
1
1
5
@BKleinstiver
This work would not have been possible without the support of
@davidwtaylorjr
, and our collaborators Kenneth Johnson and
@IlyaFinkelstein
.
@GraceHibshman
and I will be presenting this story at
@crispr2023
next week
0
0
4
Congratualtions to my co-authors
@_aparicioC
@NobregaFL
@StanBrouns
@davidwtaylorjr
. This was the first project I took on in the
@davidwtaylorjr
lab, and I'm really proud of the combination of cryo-EM, biochemistry and in vivo work we used to understand this fascinating system
1
1
5
This has been a fantastic project to work on in the
@davidwtaylorjr
team, and it's great to be able to combine structural biology, biochemical assays and in vivo work to tease apart the mechanism of this enigmatic and fascinating system.
0
1
5
Furthermore, methylation reduces the essential helicase activity of DrmAB. Work by
@_aparicioC
in parallel also showed that methylation determines plasmid conjugation efficiency.
1
0
5
@MartinPacesa
@kingcada
@lisaeshunwilson
@lindsey__young
@kylelmorris
@FrommSimon
@dabiophysicist
@BJ_Greber
There you go! Who even needs mass spec any more…
1
0
5
What a great way to celebrate
#RNAday
! First manuscript from my postdoc, determined a lovely cryo-EM reconstruction of a weird and unusual CRISPR complex
0
1
5
3.
@_aparicioC
purified a core complex containing essential factors DrmA and DrmB, and I determined structures of DrmAB bound to DNA and ADP. 3/n
1
0
5
@Seb_Fica
@BJ_Greber
@CrollTristan
@RadoDanev
@AbhayKot
If anything, you'd be able to see negative charges even more poorly in high resolution EM maps since said residues tend to have negative electron scattering factors at high spatial frequencies
2
0
5
A great opportunity at a great meeting. Thanks to
@RuedaLab
and
@NucleicAcidUK
for a wonderful day! (Also, check out my beautiful reconstruction!)
Another great talk about viral RNA chaperones delivered by
@jpkbravo
at 15th RSC Nucleic Acids Forum in London. Many thanks to
@NucleicAcidUK
and
@RuedaLab
for organising this meeting!
0
1
8
0
0
4
It was a great honour to present at such an exciting session at this wonderful meeting! Lovely feedback from some big players in the field as well!
#RNA2019
Very proud of Jack Bravo
@jpkbravo
a grad student from
@Astbury_BSL
who has just presented our work at
#RNA2019
in front of Tom Czech, Joan Steitz, Eric Westhof, Olke Uhlenbeck, David Lilley to name a few. Thanks to our brilliant collaborators
@anton_calabrese
and
@anders_barth
0
2
16
0
0
4
@rafapinilla92
That’s what we proposed for DISARM - methyltransferase and nulease modules are clearly important yet DISARM still works if they’re not present. We speculated that other host methyltransferase and PLD nucleases can compensate, making those modules redundant
1
1
4
Fantastic work! Really great to be a (tiny) part of this exciting project
0
0
4
@allostericstate
CC scores of 0.3? How can this kind of work continue to make it through peer review?
1
0
3
@ATinyGreenCell
We are working on depositing the plasmid to Addgene! For commercial applications you'd have to contact
@davidwtaylorjr
0
2
4
@Roisin_OBrien_
@davidwtaylorjr
The "more" is a big deal! Amongst the first (after Cas12a2 and new
@antigenman
structures) structures from the new Glacios scope at
@ut_mbs
@TexasScience
, the first
@davidwtaylorjr
lab in vivo assays, and the first cross-subtype Acr inhibition structure. What a killer story!
0
0
3