John Marshall
@jomarnz
Followers
274
Following
1K
Statuses
2K
Bioinformatics tools developer at large. Will edit @GA4GH specifications for food. @[email protected]
Wānaka, New Zealand
Joined January 2016
RT @GeorgeMonbiot: This is my farewell post. I am not deleting this account, as I don’t want to lose the archive, and I don’t want my name…
0
415
0
RT @howie_hua: Be on Santa's nice list by simply conveying what you mean mathematically clearly so we can stop having these PEMDAS argument…
0
335
0
@Psy_Fer_ @smllmp Mostly similarly burned in and by context of the values, but from time to time I need to type “man sam” to remind myself of the indexes of more obscure columns. (Needs a semi-well installed HTSlib with man pages included and perhaps MANPATH set if installed to a weird prefix.)
0
0
2
@camerongenomics @BonfieldJames @biomonika This sounds a lot like the functionality provided by my htsget-server-supporting branch from a long time ago, that is somewhere in the middle of my list of stuff to be dusted off someday:
0
0
1
RT @htanudisastro: 📢 New preprint alert: We’ve just published a deep dive into the role of tandem repeats (#TRs) in single-cell gene expres…
0
34
0
@yvazirabad Some indicative numbers on some oldish unaligned short read data: gzipped FASTQ 1.7G, BAM 1.5G, CRAM 1.2G Sorted by minimiser with samtools sort ‑M: BAM 1.3G, CRAM 0.86G The same data but half the storage costs? It should be a no-brainer…
0
0
1
RT @WaltersLaura: This story on @NewsroomNZ Pro ($) is your periodic reminder that our PM writes all emails in blue comic sans https://t.c…
0
54
0
@luka_culibrk I did wonder how you'd managed to avoid being exposed to hex notation 🤣 Always happy to have an excuse to publicise useful tidbits like `samtools flags` though!
1
0
0
The “just use conda-forge & bioconda” answers to the current conda kerfuffle mostly gloss over the fact that those channels' artefact repository infrastructure appears to be owned and operated by Anaconda. Good to see that (and a path to an alternative) discussed in this posting.
@NM_Reid Hey @NM_Reid – for the open source (not "defaults" channels) like @condaforge and bioconda we have worked hard to ensure that there is no vendor lock in. They remain _free to use_.
0
0
1
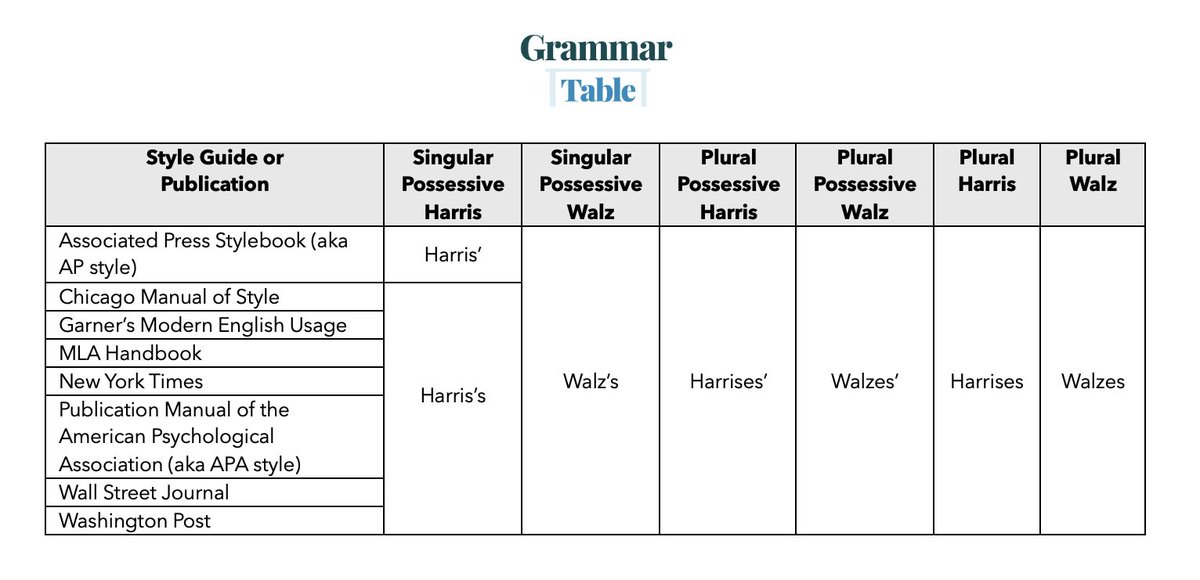
RT @GrammarTable: I'm seeing mistakes on the internet. I hope this Grammar Table table helps.
0
112
0