wei yang
@VinetteY
Followers
171
Following
284
Statuses
27
PhD student @uwgenome | @JShendure lab| single cell genomics, developmental biology | she/her
Seattle, WA
Joined June 2018
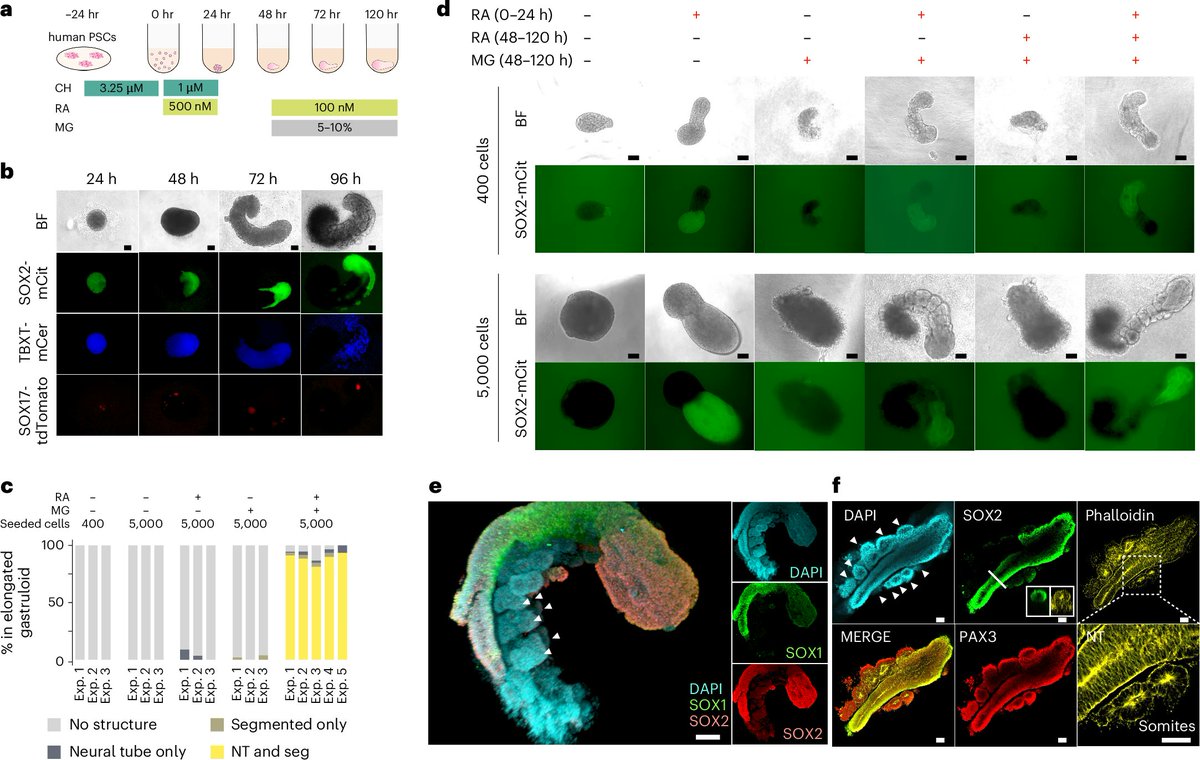
I am so thrilled to share our work of human RA gastruloid finally out on @NatureCellBio. It has been a wonderful journey to work with a fantastic stem cell scientist @Nobu_Hamazaki and many talented collaborators in @JShendure lab. Excited to contribute to the field of stembryo!
Excited to share our human RA-gastruloid model in @NatureCellBio! Early retinoic acid supplementation, followed by later Matrigel addition, robustly induces human embryonic morphologies and diverse cell lineages, including somitic, renal, cardiac, and neural cell types in #gastruloid.
0
4
22
RT @jb_lalanne: After a constructive set of reviews, our single-cell reporter work is OUT Check out the briefing if…
0
56
0
RT @Yi__Fu: Excited to share my PhD work in @Agnelsfeir’s lab describing a method to engineer mitochondrial DNA deletions in human cells an…
0
18
0
RT @TroyMcDiarmid: Beyond stoked to share our latest, entitled “Diversified, miniaturized and ancestral parts for mammalian genome engineer…
0
14
0
RT @FloChardon: Genomic tech dev is my favorite area of science, and today our paper describing our multiplex CRISPRa screening method to i…
0
26
0
RT @riddhimankg: Very happy to share our latest w/ @dschweppe1 and @Nobu_Hamazaki labs, where we map the temporal dynamics and proteomic la…
0
20
0
RT @wchenomics: Super excited to share that ENGRAM is out today! ENGRAM cells are programmed to write their histories into the genome, reco…
0
130
0
RT @XiaoyiLi10: Our work on studying chromatin context-dependent regulation of prime editing is officially out on @Cell.
0
72
0
RT @JShendure: Our latest out today in @Nature. We profiled 12 million single cells from mouse embryos spanning gastrulation to birth, defi…
0
162
0
RT @Nobu_Hamazaki: Congrats,@CXchengxiangQIU, @bethkarenmartin, Ian, and @JShendure! What a fantastic paper which would be a long-years bas…
0
9
0
RT @JShendure: Advanced human gastruloids, our latest preprint from the brilliant @Nobu_Hamazaki ( & @VinetteY, rob…
0
37
0
RT @Nobu_Hamazaki: We (with @VinetteY and @JShendure) are thrilled to announce our new paper on the "human RA-gastruloid" model, showcasing…
0
66
0
RT @Nobu_Hamazaki: 🎉Exciting news! Starting this summer, we're launching a groundbreaking lab at @UW! Unraveling & reconstructing mammalian…
0
20
0
RT @XiaoyiLi10: Thrilled to share my work describing chromatin-context dependent regulation of prime editing and our effort in leveraging t…
0
53
0
RT @JShendure: Today we release in @biorxivpreprint a pair of related methods, DNA Ticker Tape & ENGRAM, for multiplex, temporally ordered…
0
274
0
RT @_Choi_Junhong: At @JShendure lab, we found yet another use of prime editing – precise genome deletion! Building on the prime editing de…
0
24
0
RT @junyue_cao: Please check our newest work! A human cell atlas of both gene expression and chromatin accessibility across 15 fetal tissue…
0
39
0
RT @diegoisworking: Thrilled to share what I’ve been working on during the first year of my postdoc — a powerful experimental design to fig…
0
74
0
So excited to give my first conference talk at @acm_bcb tomorrow on my PhD rotation project with @thabangh and @jbilmes on the application of submodular maximization to summarizing single-cell RNA-seq data😎And thanks a lot for @jmschreiber91 ‘s cool software apricot!
0
3
16
WE NEED TO KNOW!!
Does anyone know why @illumina's P5/P7 sequences are named "5" and "7"? I can guess that the "P" stands for primer, but why those numbers? I've spent the last hour in a rabbit hole and have asked our NGS experts here at @10xGenomics, but no one knows!
0
0
1