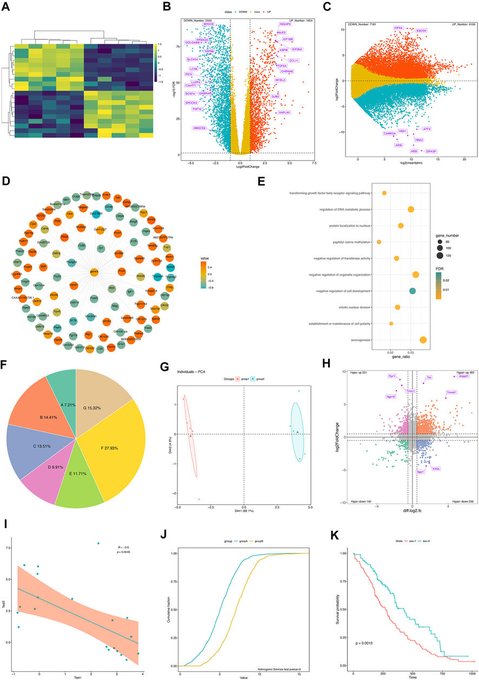
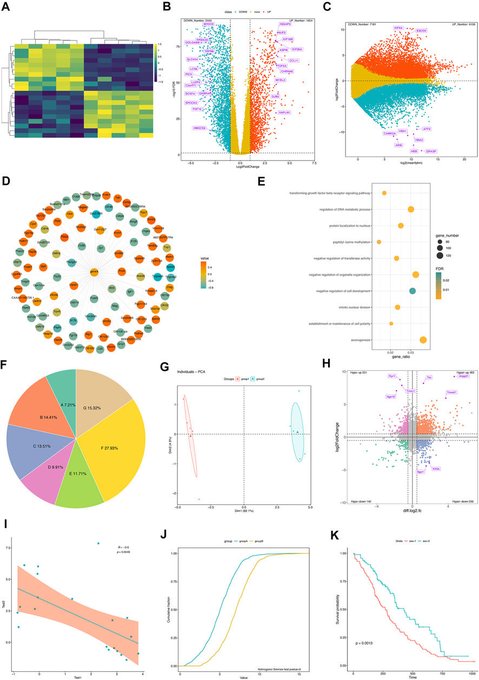
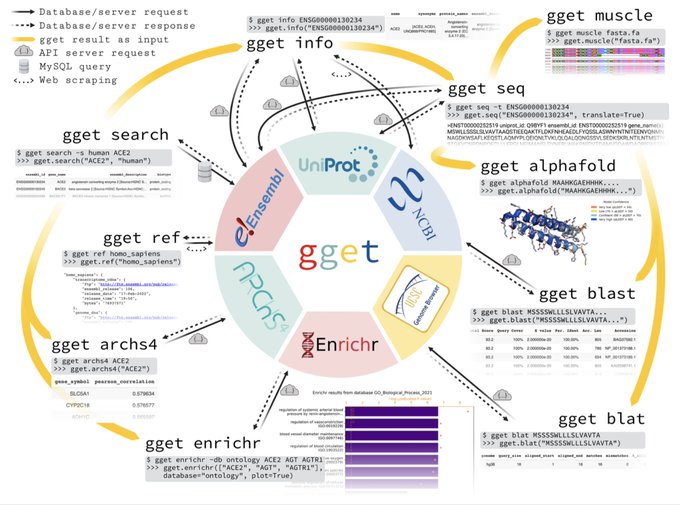
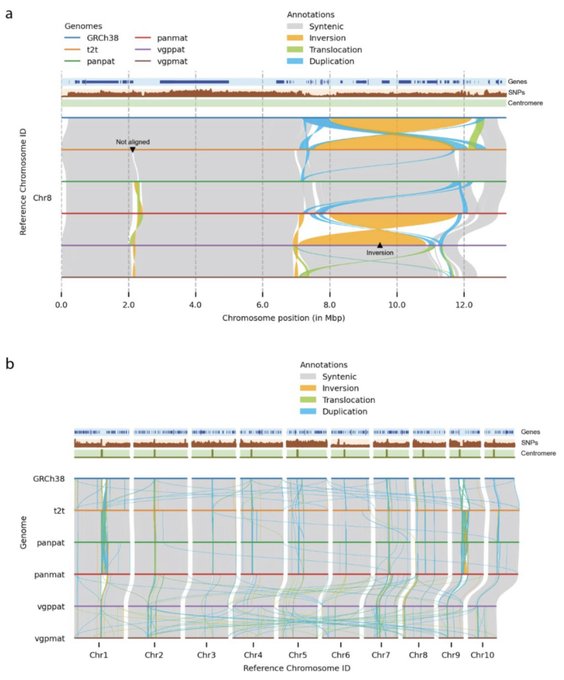
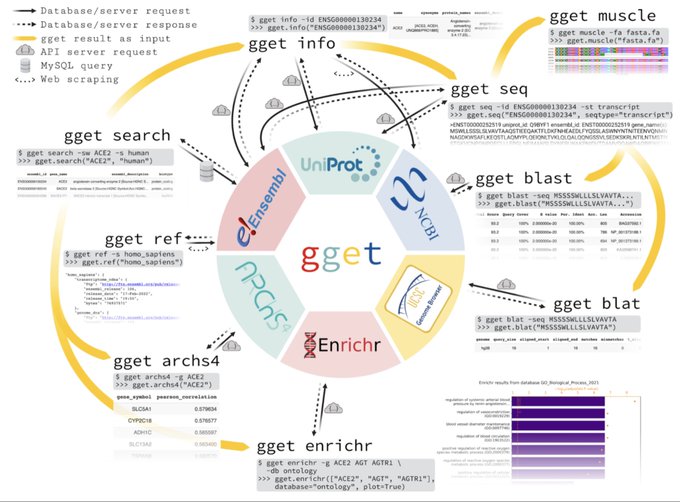
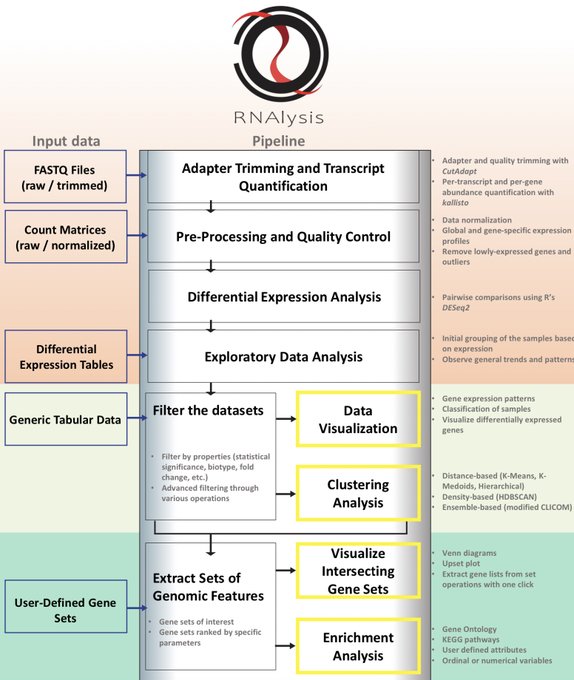
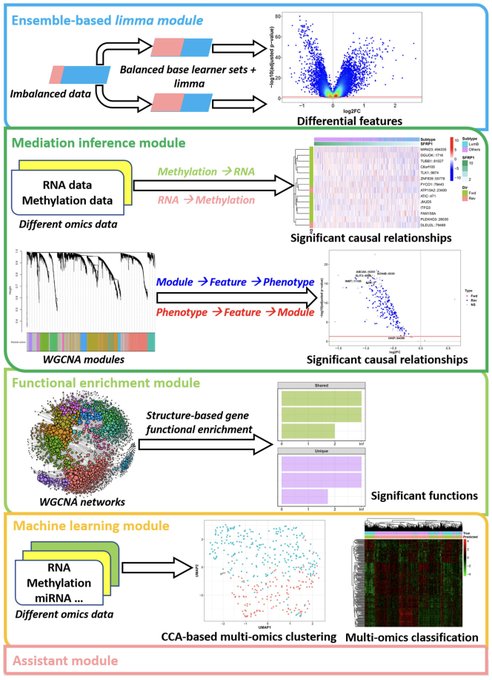
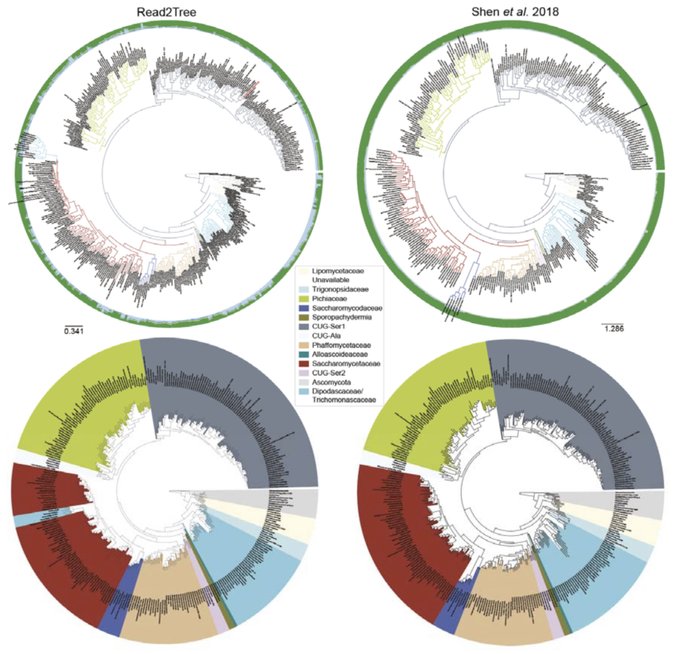
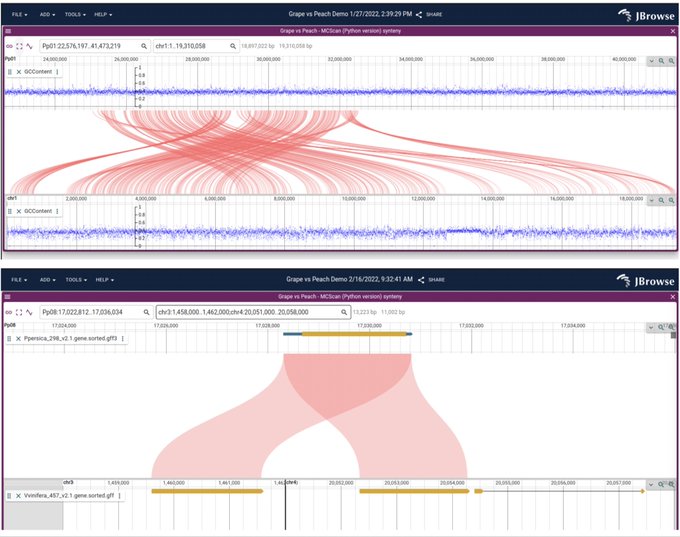
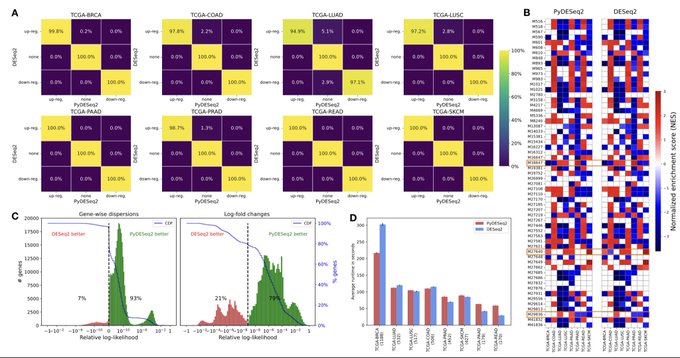
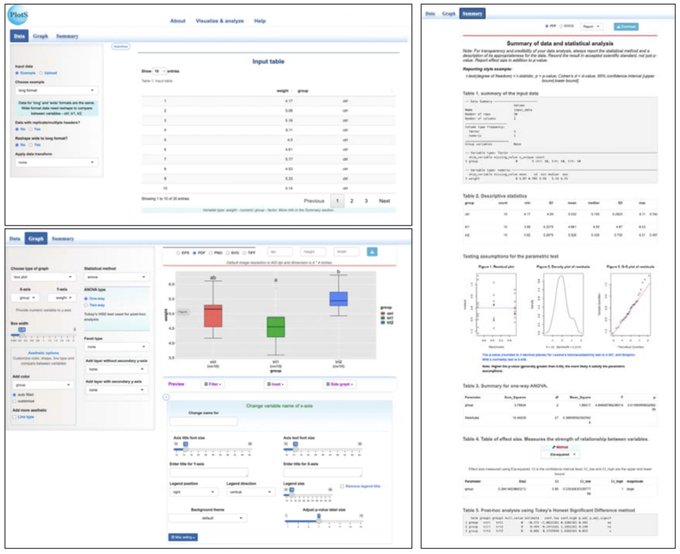
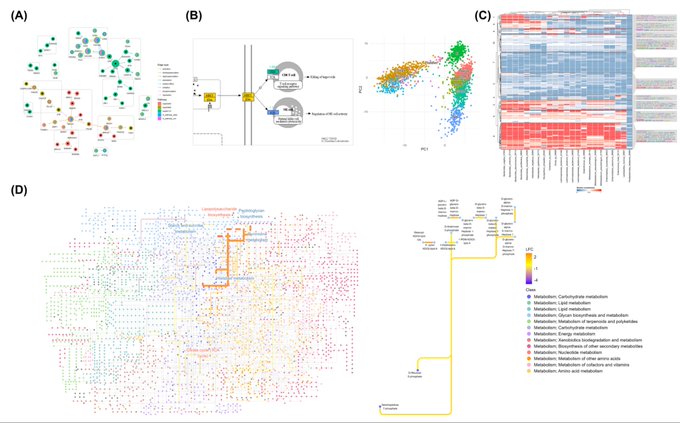
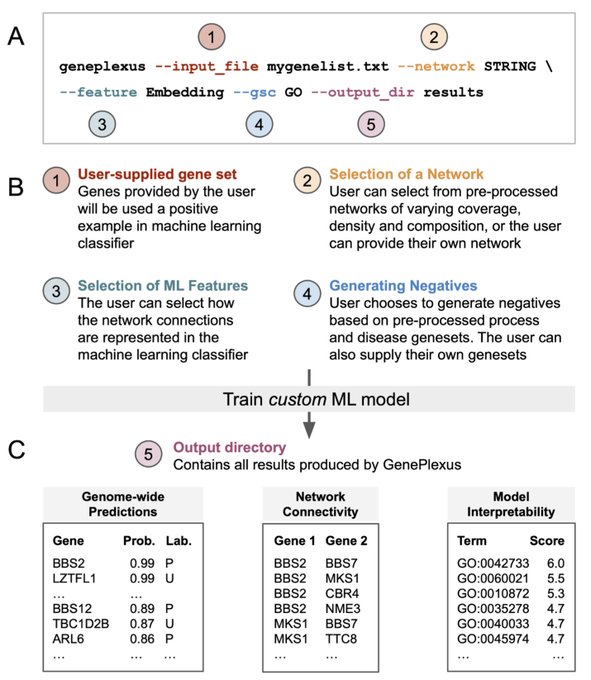
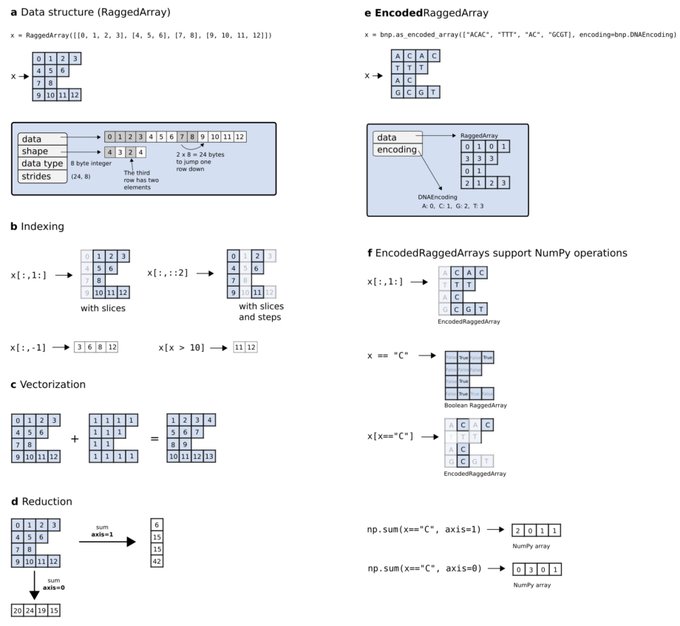
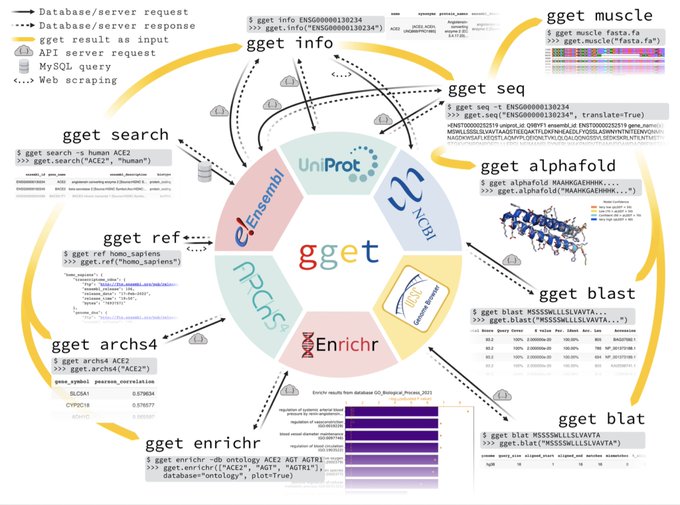
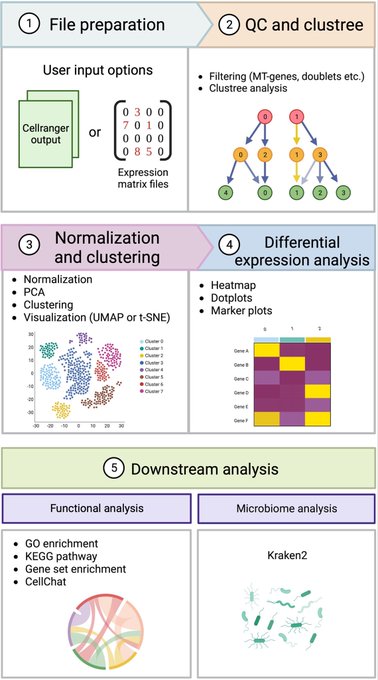
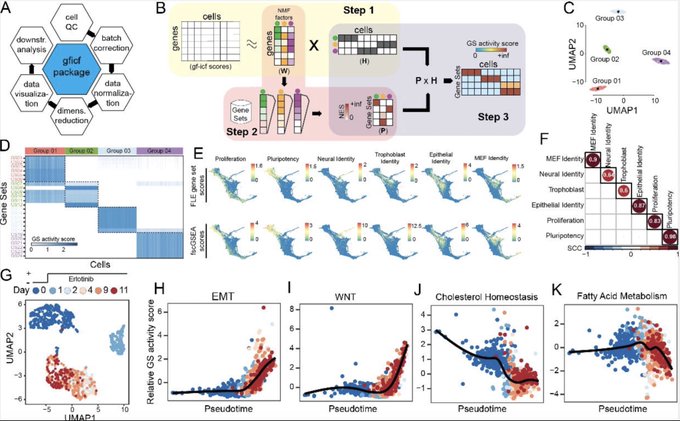
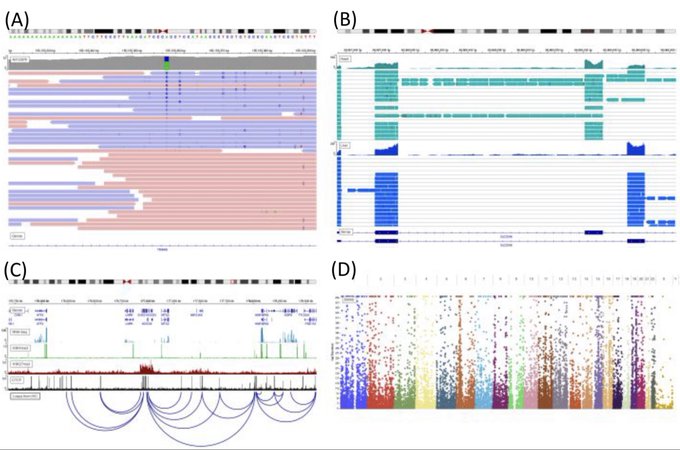
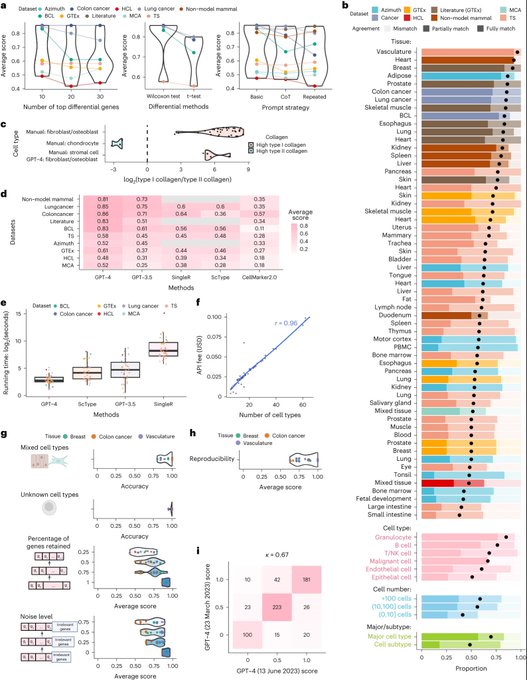
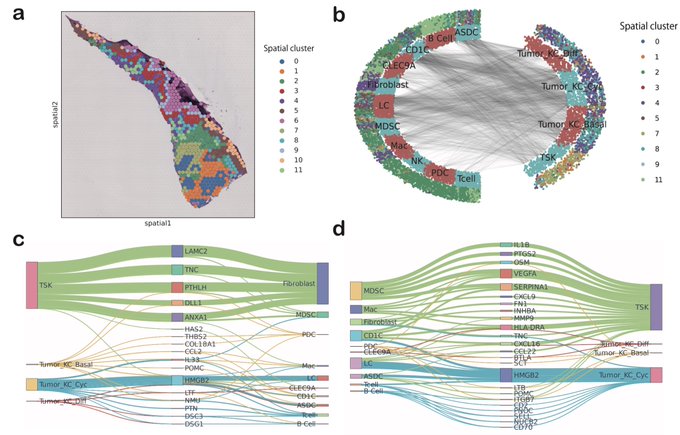
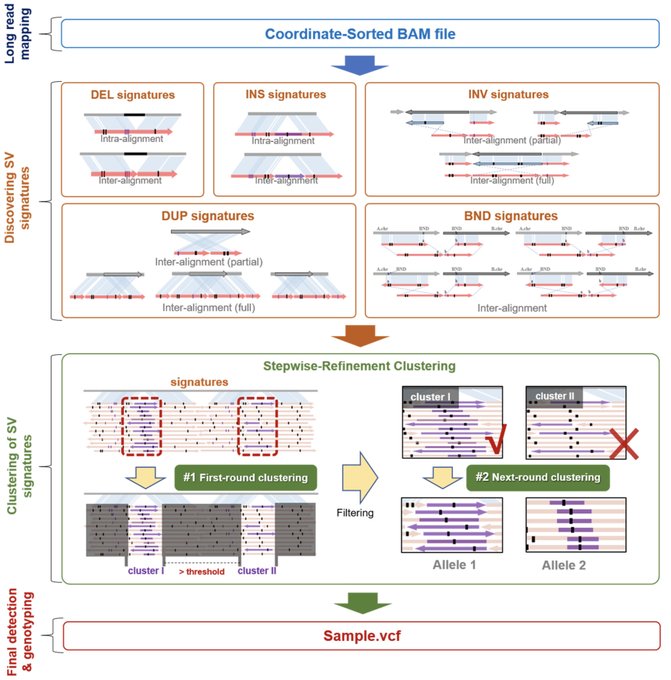
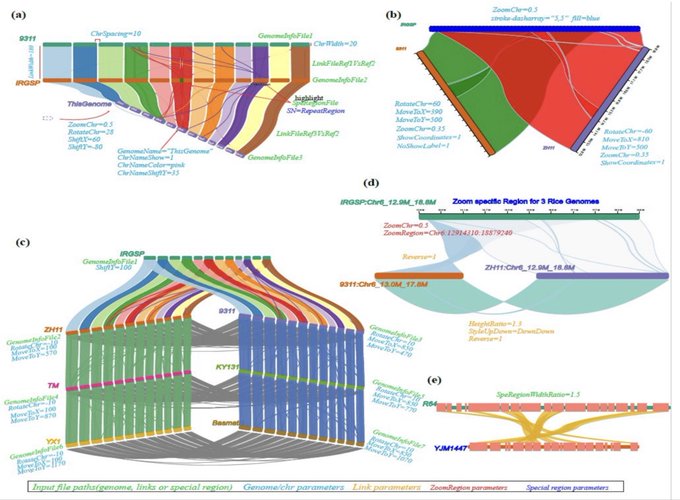
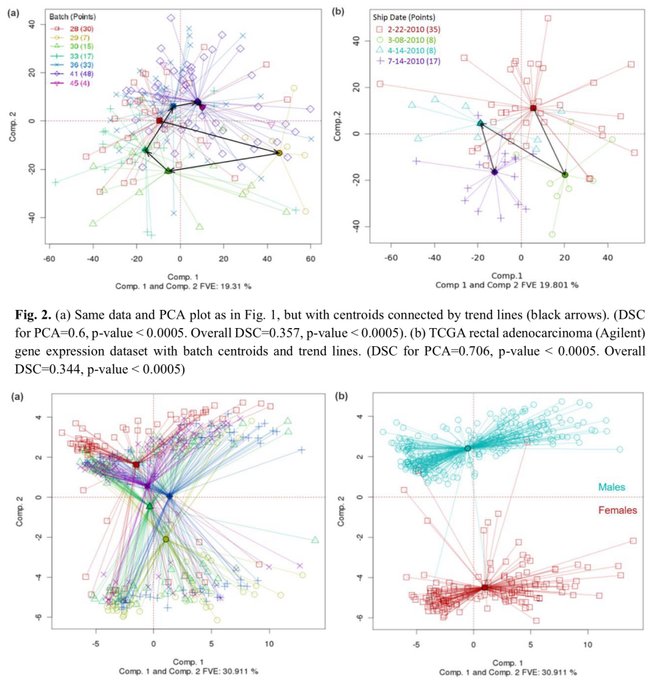
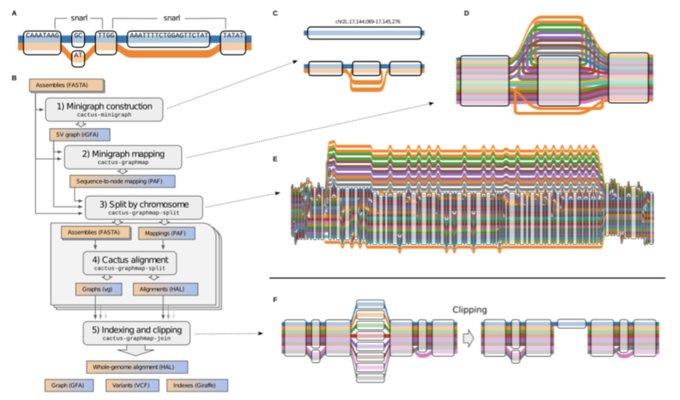
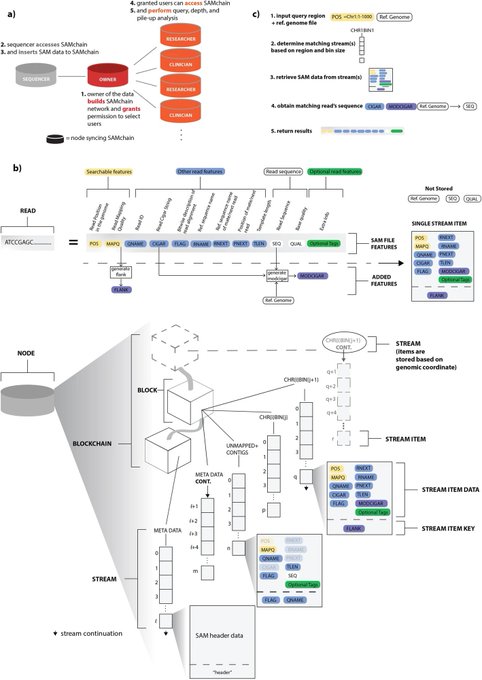
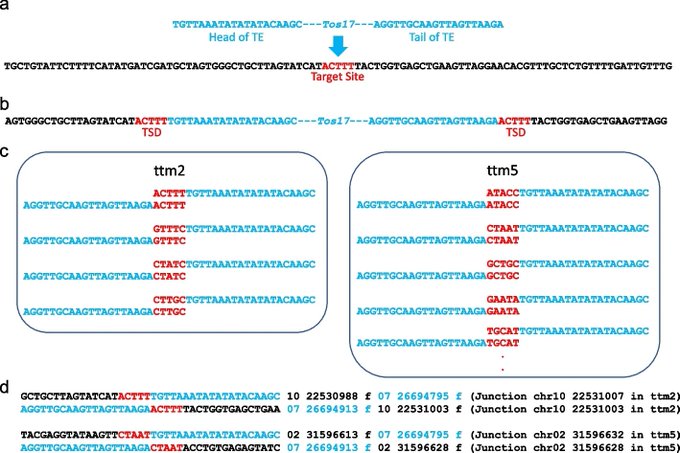
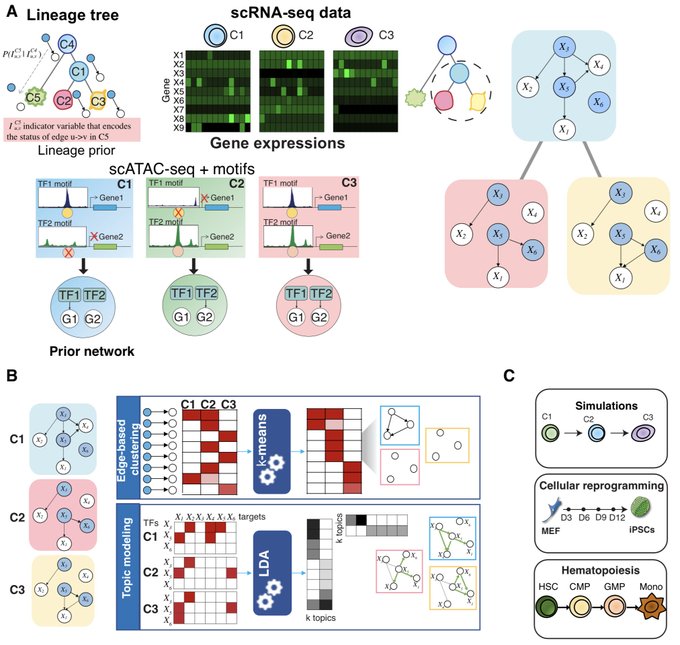
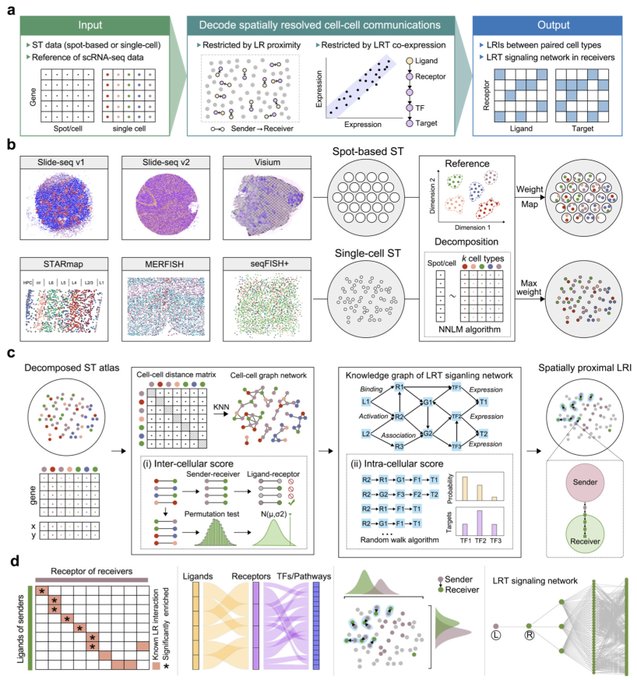
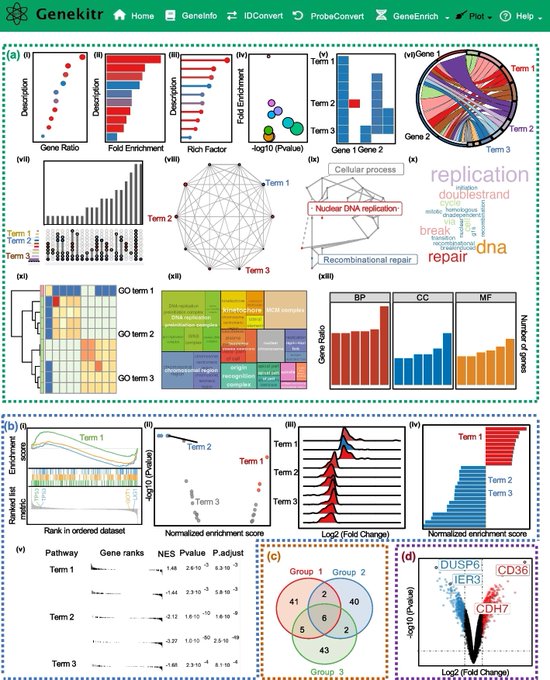
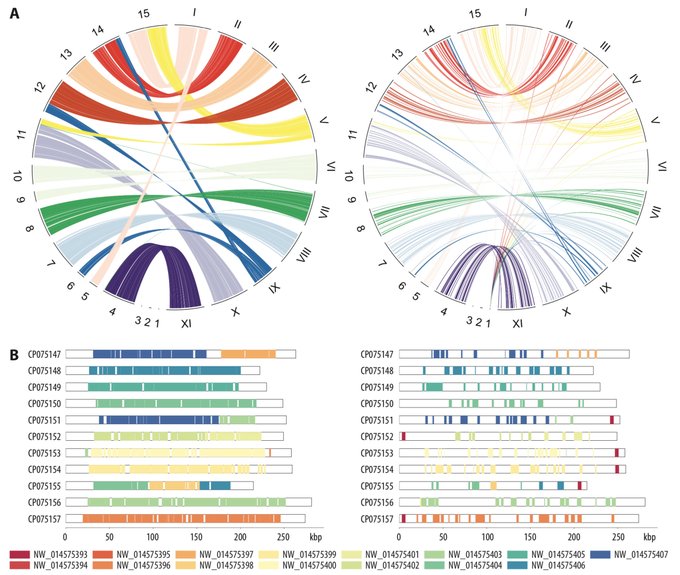
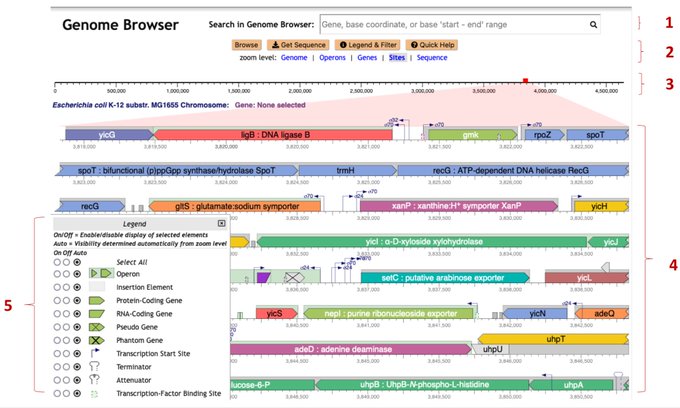
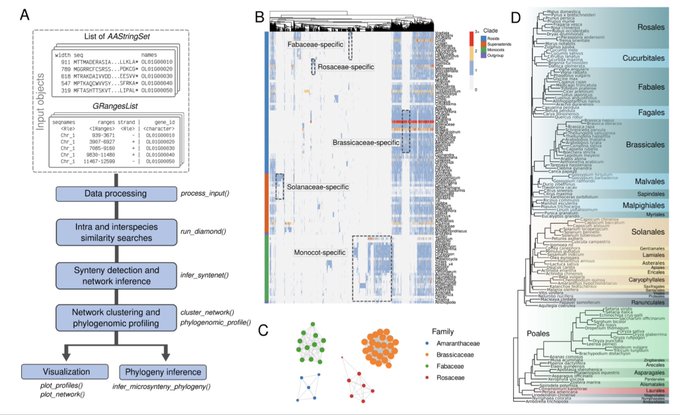
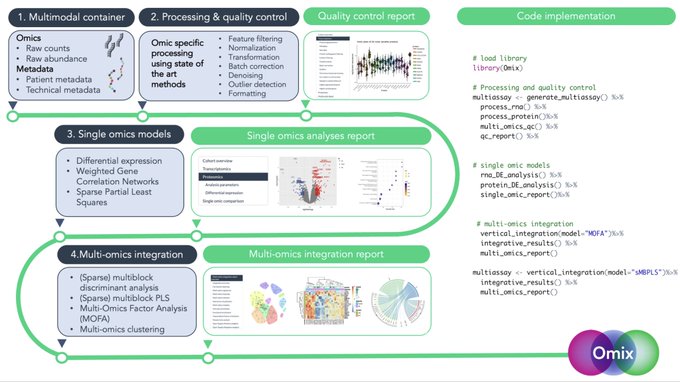
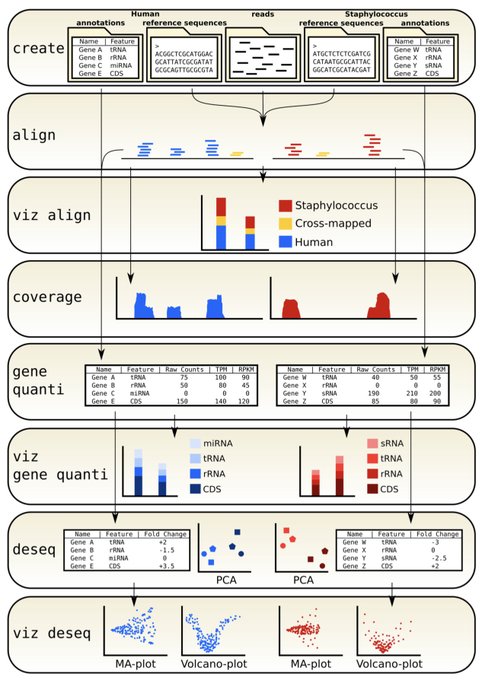
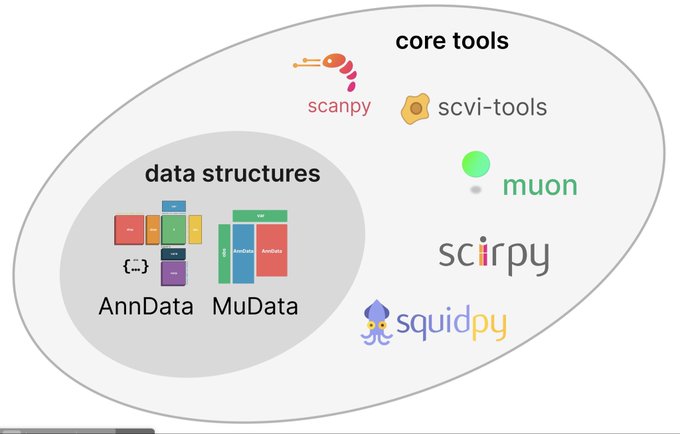
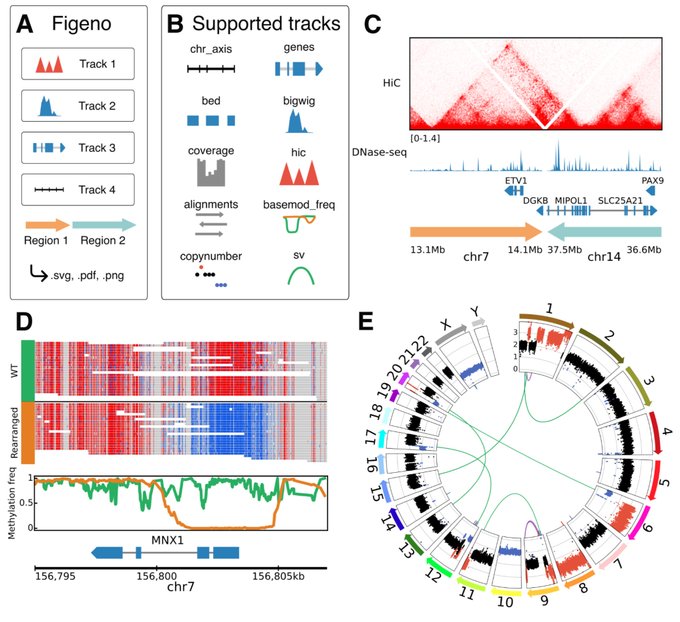
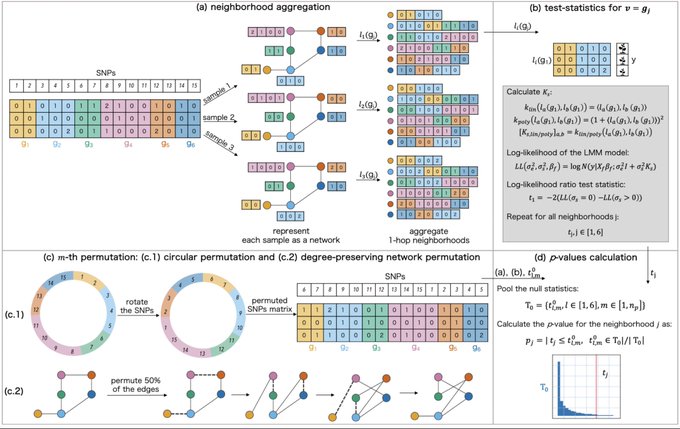
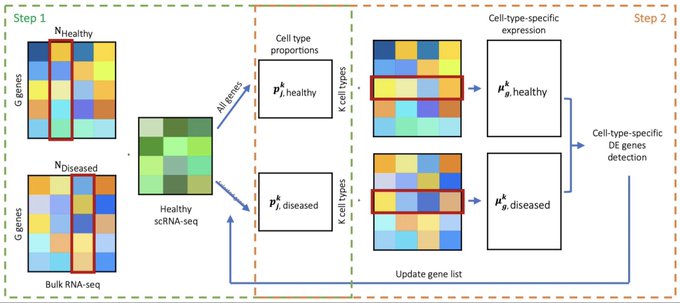
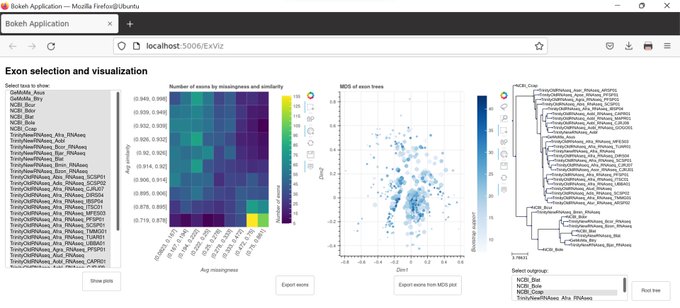
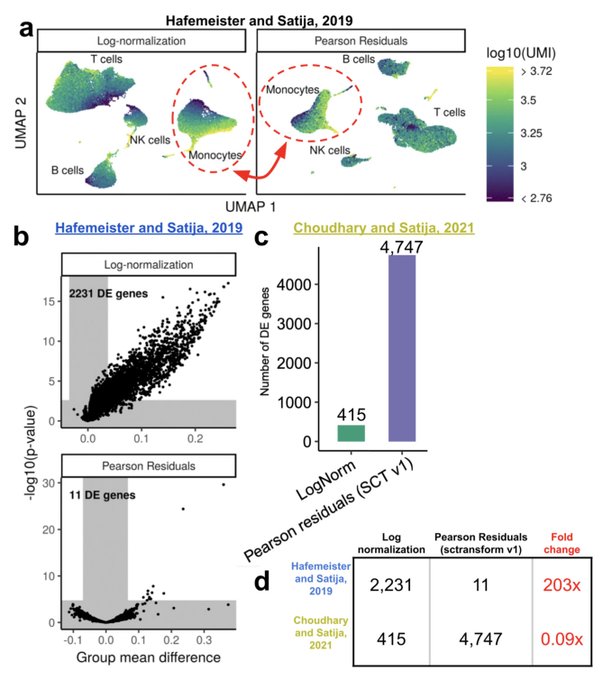
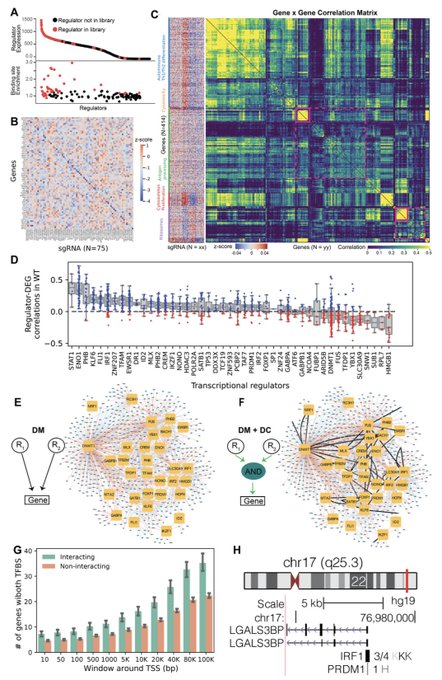
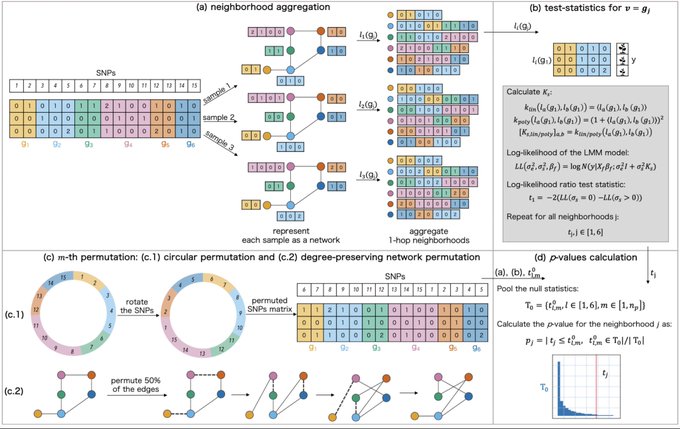
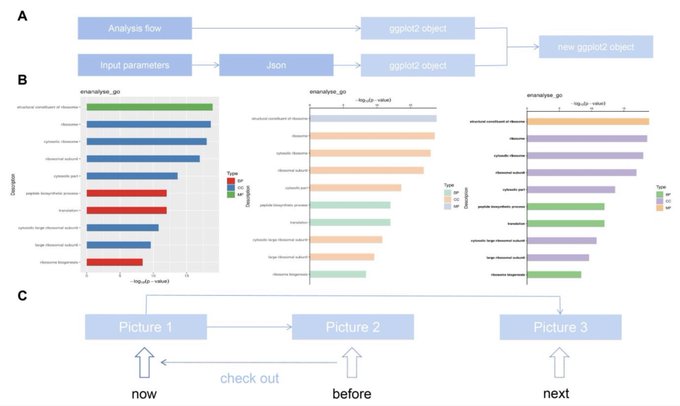
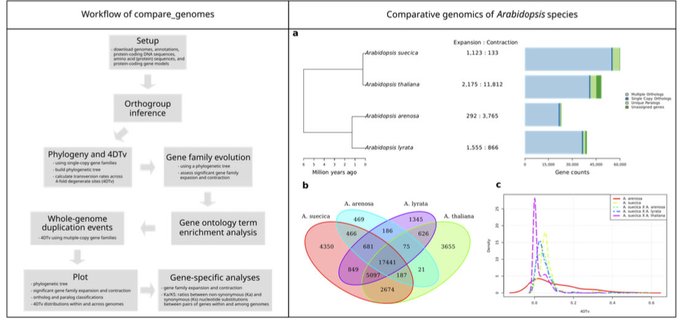
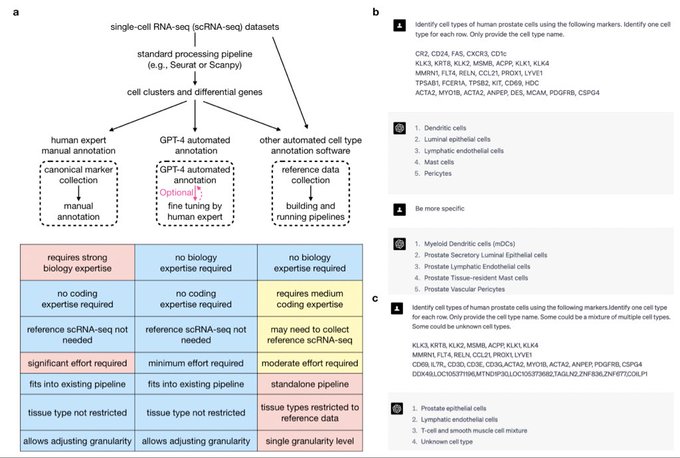
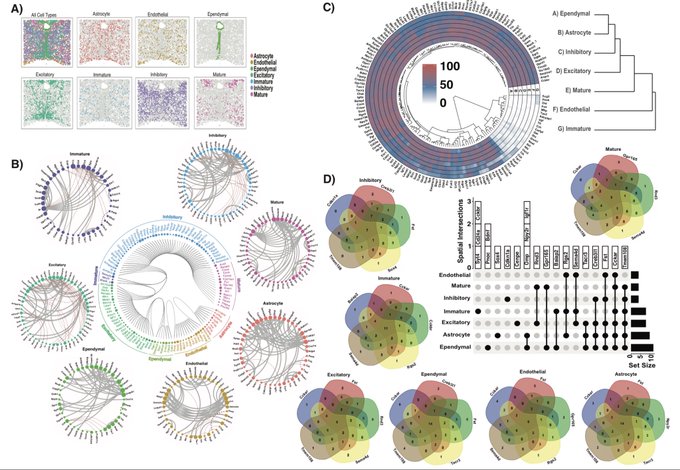
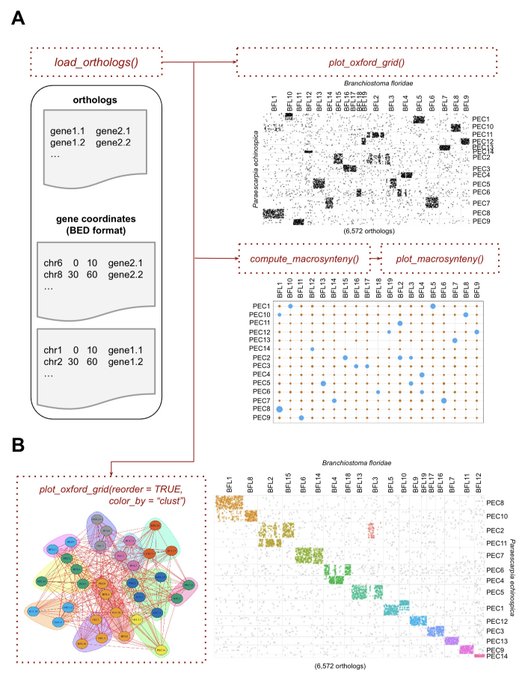
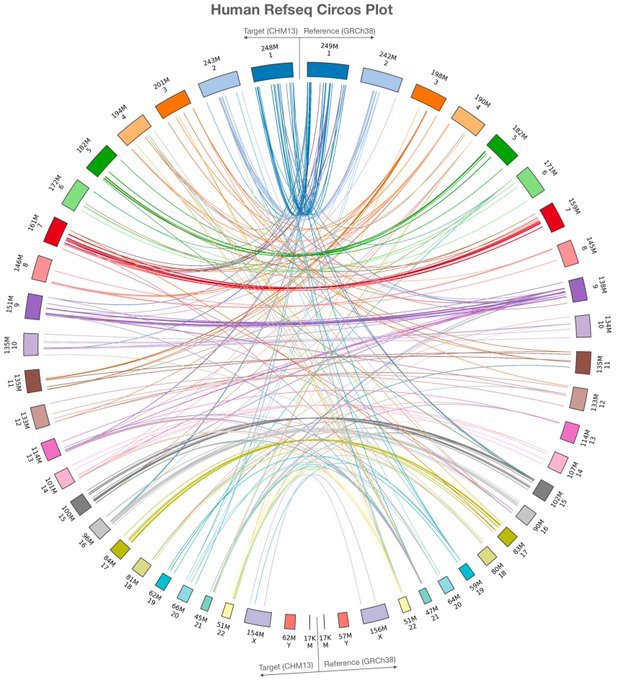
antisense.
@razoralign
Followers
14,196
Following
67
Media
13,848
Statuses
84,728
God made everything out of nothing. But the nothingness shows through.
Joined November 2009
Don't wanna be here?
Send us removal request.
Explore trending content on Musk Viewer
Adalet
• 242820 Tweets
Chester
• 112909 Tweets
ベイマックス
• 109370 Tweets
Merchan
• 102490 Tweets
Jean Carroll
• 81133 Tweets
Milli
• 76841 Tweets
#2KDay
• 76197 Tweets
Eagles
• 68396 Tweets
Ayşenur Ezgi Eygi
• 40881 Tweets
Franco Escamilla
• 40443 Tweets
#CristinaMostraElTitulo
• 36548 Tweets
Packers
• 31129 Tweets
Barcola
• 31055 Tweets
Dick Cheney
• 22786 Tweets
Mert
• 16855 Tweets
Bafana Bafana
• 15974 Tweets
Ganesh Chaturthi
• 15542 Tweets
Galler - Türkiye
• 14877 Tweets
Kenan
• 13602 Tweets
#FRAITA
• 12925 Tweets
Chaine
• 12597 Tweets
Go Birds
• 12344 Tweets
Sergio Mendes
• 11027 Tweets
#BizimÇocuklar
• 10994 Tweets
Olise
• 10126 Tweets
Last Seen Profiles