josephblayney
@josephblayney
Followers
94
Following
101
Media
0
Statuses
28
PhD gene regulation 🧬🧫 • food, wine, books, films & weightlifting 🍔🍷📖🎞🏋️ • @UniofOxford • VMO
Oxford, England
Joined October 2019
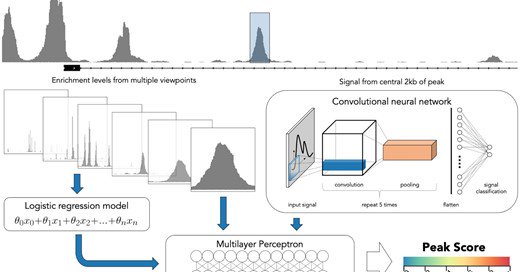
Our machine learning approach to peak calling "LanceOtron: a deep learning peak caller for genome sequencing experiments" has been published. If you want better peak calls and less false positives in your ChIP-Seq results, please give it a try!
academic.oup.com
AbstractMotivation. Genome sequencing experiments have revolutionized molecular biology by allowing researchers to identify important DNA-encoded elements
1
9
40
#Superenhancers function in an orientation dependent manner,challenging #enhancerbiology paradigm. @Hele_Fran @josephblayney @MariaC_Su @CazzHarrold @jrmmhughes @DownesDamien @MariekeOudelaar @NatureGenet @Hannah_K_Long @jojdavies @generegulation @MRC_WIMM
https://t.co/Dtd8ZLW3VY
biorxiv.org
Transcriptional enhancers regulate gene expression in a developmental-stage and cell-specific manner. They were originally defined as individual regulatory elements that activate expression regardl...
1
19
48
Only one week left to apply for our postdoc position! If you're interested in human genetics, chromatin, and gene regulation then check it out! Informal inquiries always welcome
🚨Job Alert🚨 We’re hiring a postdoc to work on genetic diseases of chromatin! 🧬 🏫Come work with us at @HumanGeneticsOx in the beautiful @UniofOxford 🗓Closing 6 July 📨 More details at https://t.co/TRLXH3zl7q Pls share and RT! #PostdocJobs #GeneticsJobs #genomics #cohesin
0
10
4
Have you ever made backwards DNA??? Brendan did - on purpose!! . Check out what happens to "vanilla" DNA in yeast and mouse cells. What do they do with DNA they've never seen before? #darkmatterproject strikes again ... courtesy of @bcamellato
biorxiv.org
Up to 93% of the human genome may show evidence of transcription, yet annotated transcripts account for less than 5%. It is unclear what makes up this major discrepancy, and to what extent the excess...
1
6
29
0
0
1
Lastly, facilitators show functional hierarchy, although this is encoded in their relative positioning rather than their sequences. (6/6)
1
0
1
Without facilitators, the classical enhancers exhibit reduced mediator recruitment, eRNA transcription and enhancer-promoter interactions. (5/6)
1
0
2
Surprisingly, the two strong enhancers are unable to activate high levels of gene expression by themselves. They rely on the three non-enhancer, “facilitator” elements to facilitate their functions. (4/6)
1
0
2
The α-globin SE contains 2 strong classical enhancers and 3 elements bearing the bioinformatic signature of enhancers, but with no apparent enhancer activity. (3/6)
1
0
2
Through comprehensively rebuilding the endogenous α-globin super-enhancer (SE), we show that SEs comprise functionally distinct constituent elements…classical enhancers, and novel “facilitators” which augment enhancer activity. (2/6)
1
0
1
Very excited to share our new pre-print!: Super-enhancers require a combination of classical enhancers and novel facilitator elements to drive high levels of gene expression https://t.co/01PPmH9GXM ...short thread below (1/6)
biorxiv.org
Super-enhancers (SEs) are a class of compound regulatory elements which control expression of key cell-identity genes. It remains unclear whether they are simply clusters of independent classical...
5
27
147
🚨Job Alert🚨 We’re hiring a postdoc to work on genetic diseases of chromatin! 🧬 🏫Come work with us at @HumanGeneticsOx in the beautiful @UniofOxford 🗓Closing 6 July 📨 More details at https://t.co/TRLXH3zl7q Pls share and RT! #PostdocJobs #GeneticsJobs #genomics #cohesin
3
36
47
Kicking off 2022 with my first senior #author research paper.The novelty lies in reimagining the use of a well-established #mESC #invitro system harnessing new technologies to serve genetic studies @Hele_Fran @RobBeagrie @DM_Jeziorska @MRC_WIMM @PLOSONE
https://t.co/oBWshTDdFf
journals.plos.org
Mouse embryonic stem cells (mESCs) can be manipulated in vitro to recapitulate the process of erythropoiesis, during which multipotent cells undergo lineage specification, differentiation and...
12
9
44
Really happy to see our latest work out on bioRxiv. We used live-cell antibody staining to facilitate the study of alpha-globin transcriptional bursting dynamics throughout erythropoiesis and identified changes in transcriptional noise during differentiation. Check it out! ⬇️⬇️⬇️
On-microscope staging of live cells reveals changes in the dynamics of transcriptional bursting during differentiation https://t.co/fsy8U6n5U2
#bioRxiv
0
8
19
Delighted to share my lab’s @jdavieslab first big paper in @Nature! We’ve developed a new 3C technique called Micro Capture-C that can generate base pair resolution 3C data https://t.co/Mgr26d2bTE
35
260
819
Why are ultraconserved enhancer sequences ultraconserved? Our latest attempt to solve the mystery, led by @Pribaikalochka Here is the TL;DR thread (1/n) https://t.co/27AsCzOD0z
5
121
304
Enhancers may cooperate synergistically to determine cell fate. Synergistic enhancers are defined by eRNA synthesis, depend on cell-specific TFs and differ form occupancy-defined superenhancers. Great collaboration with Graf & Söding labs. Finally out: https://t.co/en8ctYctzz
0
47
203
Out Today! Using our new method (scaRNA-seq) we found enhancers regulate PolII Recruitment and Initiation – rather than Pausing @MolecularCell @generegulation
#GeneRegulation
https://t.co/5EdBrKaEkk
1
47
156
I’m very happy to share this @NatureRevGenet review in which Doug Higgs and I discuss the most recent insights into genome organisation, gene regulation and their relationship: https://t.co/H19OMV8qyv
#3Dgenome #generegulation
0
100
345
Many insights into gene regulation result from in-depth characterisation of model gene loci. We reviewed recent studies of the alpha-globin cluster and lesson learned about gene regulation & genome organisation: https://t.co/KT1ZNViufn, with @RobBeagrie, @MiraKassouf & Doug Higgs
1
9
39