Charles Breeze
@charles_breeze
Followers
419
Following
346
Statuses
483
Researcher in computational genomics and molecular epidemiology @theNCI | Hon. Assist. Professor @UCL | Prev @UniofOxford, @emblebi, Altius | Opinions mine
Bethesda, MD
Joined June 2014
RT @POPGEM_Lab: 🚀New paper by @Madhur_B_Singh on smoking & DNA methylation via twin-based MR using @NTRscience data! Supervised by @mcneale…
0
4
0
RT @doctorveera: A thread on some of the fascinating noncoding genetic discoveries from 2024. I've written long form posts and discussed on…
0
38
0
RT @jenny_vandongen: New paper by our PhD student Austin van Asselt @NTRscience Epigenetic signatures of asthma: a comprehensive study of…
0
1
0
Fantastic talk by @cardenaasca in the @illumina Education Session on molecular markers of aging and the need for increasing the #diversity of participants in #epigenetic studies 🤩🧬 #ASHG24
1
2
8
RT @JoshTycko: Our new paper on epigenome editing is out! We mapped which effector domains regulate transcription across genomic, cell ty…
0
73
0
RT @jenny_vandongen: Check out our new paper: Examining the Utility of the Mammalian Methylation Array for Pan-Mammalian Analysis of Monozy…
0
2
0
My first career podcast interview! Couldn't be happier! 🎉🎉🎉🎉 Thank you @EverythingEpi @TruDiagnostic ! I would like to thank @NCIEpiTraining, @jloukissas, @NoraFranceschi2 and @mitchiela for support and advice! #KidneyDisease #epigenetics
0
5
13
RT @maxewilkinson: My favourite discovery ever has just come online. Can I please tell you about some seriously wacky molecular biology? Th…
0
1K
0
@tangming2005 Nice list! One tool I might add is FORGEdb, which integrates data across TFs, enhancers and histone modifications #ENCODE4
🧬 Our #ENCODE4 paper is out now @GenomeBiology! Dissecting uncharted regions in our DNA and their influence on our health. 🧬 Dive into the world of disease genetics and genomic "dark matter" with us as we explore our trailblazing new method. Ready? Let’s go! 🚀 🧵 #Genomics #GenomeFunction #ScienceTwitter 1/
0
2
3
RT @Dr_James_Lee: There's been amazing coverage of our new paper @Nature but I wanted to put together a walk-throug…
0
231
0
RT @EricTopol: Discovery of a major underpinning of inflammatory bowel disease in a gene desert @Nature @TheCrick h…
0
168
0
@sinabooeshaghi @anshulkundaje Nice tool! Btw if you are interested check out the code and webpage for Author Arranger: and
1
0
0
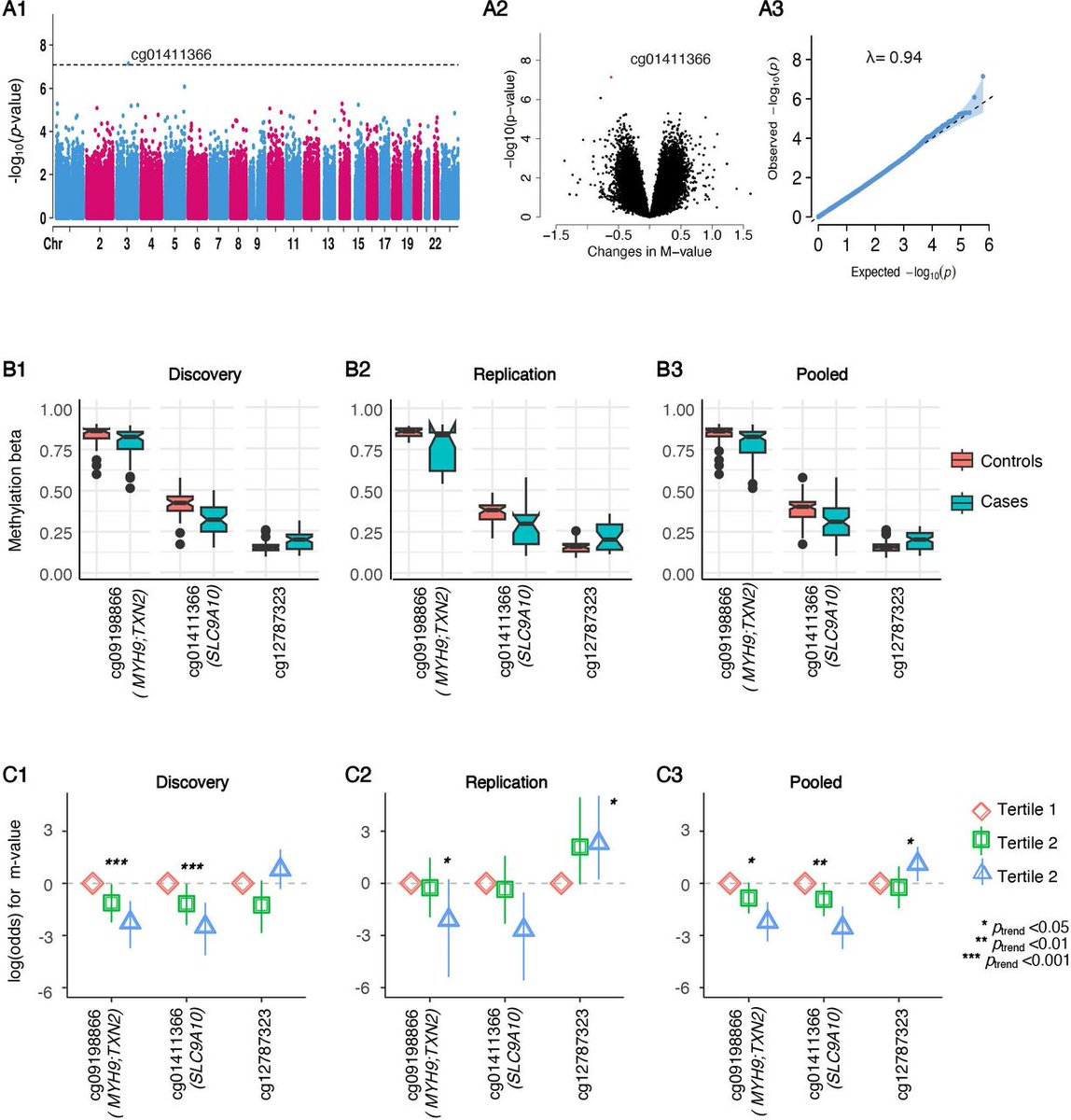
🧬 Our paper is out now @BioMedCentral! Exploring the Mysteries of APOL1-associated kidney disease through DNA Methylation 🧬 Background One of the Kahun Papyri from ancient Egypt (2nd millennium BC) describes a disease of cattle characterised by severe wasting and lethargy. The origin of the disease was a mystery. During the Middle Kingdom (2000 BC – 1300 BC) the stream course of the Nile River was adjusted and the disease became less common. The Veterinary Papyrus (from the Kahun Papyri) Further south, and a few centuries later, an acount from an explorer in Lake Malawi details: "A swelling appeared under the cow's jaw. The animal continued to graze, but soon began to show signs of emaciation, with its muscles becoming noticeably flaccid. After several months, the animal, unable to graze any longer, succumbed to extreme exhaustion and died." An example of a diseased steer At the turn of the 20th century, a devastating outbreak in humans was proof that this disease was not just limited to domestic animals. Indeed, this disease has been with humanity for such a long time that it has affected human evolution. Any form of resistance, however costly, was conferred an evolutionary advantage and became more common in affected areas. Indeed, one of the few documented gain-of-function mutations in humans ocurred in response to this disease (African sleeping sickness). An evolutionary arms race that has had repercussions to this day. APOL1: a double-edged sword Certain variants in the APOL1 gene provide gain-of-function mutations that can result in the death of human-affecting Trypanosome parasites (that cause African sleeping sickness). APOL1 kills trypanosomes by forming pores in the trypanosome lysosomal membrane. When the gain-of-function G1 and G2 variants are present, it can kill human-affecting trypanosomes. However, these variants come at a cost: high rates of kidney disease. G1 and G2 variants are associated with increased kidney disease risk and partly explain incrased kidney disease in populations of African ancestry. One enduring mystery is that not all G1 and G2 carriers get kidney disease, which seems to occur in only a subset of individuals. To better understand why not all G1 and G2 carriers get kidney disease, we performed a study to characterise the epigenetic context surrounding these disease-associated variants. We focused specifically on DNA methylation, an epigenetic mark, typically at CpG sites, that controls gene expression and has been associated with a number of different diseases, including kidney function. Better understanding of the epigenetic marks surrounding G1 and G2 may help us understand why only a subset of individuals get kidney disease. What did we find? In our paper, we performed DNA methylation analysis of 611 African American participants that had APOL1 risk genotypes, to detect differentially methylated positions (DMPs). We found DNA methylation at five CpGs was significantly associated with APOL1 risk alleles in discovery and replication studies, and one CpG-APOL1 association was independent of other genomic variants. Manhattan plot with top associated CpGs These DMPs, the first epigenetic differences associated with disease risk variants, are located close to the APOL1 gene. Specifically, the five variants are located within 60kb of the APOL1 gene (shown below). In particular, cg21092464 was associated with G1/G2 risk variants and was not found to be associated with any other genomic variant in the region. These findings, taken together with gene expression work (, advances research to better understand differences in the epigenetic landscape associated with APOL1 risk variants in individuals of African descent. The proximal epigenetic landscape of APOL1 risk variants END Huge thanks to @NoraFranceschi2, Briget Lin, and Cheryl Winkler (NCI CCR) for supporting this work! References
0
4
4
Thank you @ThoraxBMJ for highlighting our #EWAS on lung cancer in non-smokers! 🎉🎉🎉🎉
Epigenome-wide association study of lung cancer among never smokers in two prospective cohorts in Shanghai, China @MohammadLRahma2
0
2
7
10/ Huge kudos to @MohammadLRahma2 @jasonyywong @batelblechter @cardenaasca Nat Rothman, Qing Lan and other-co authors! Huge thanks also to David Christiani @DChristianiMD for his excellent commentary and @NCIEpiTraining @NCIChanock for funding and support!
0
2
2
7/ These findings underscore the significance of investigating non-smoking populations to elucidate the diverse etiological factors contributing to lung cancer. Moreover, the utilization of oral rinse samples offers a non-invasive and accessible approach to identify novel biomarkers for risk assessment and early detection.
1
1
1