Biotite
@biotite_python
Followers
252
Following
3
Statuses
114
This Python package is your Swiss army knife for bioinformatics. It allows analysis of sequence and biomolecular structure data in an easy and efficient manner.
Joined November 2017
@AminSagar1 @RDKit_org The cyclotide structure and its coordinates originate from a NMR structure ( in the example above, so in this case there is no problem with a generated conformation.
0
0
1
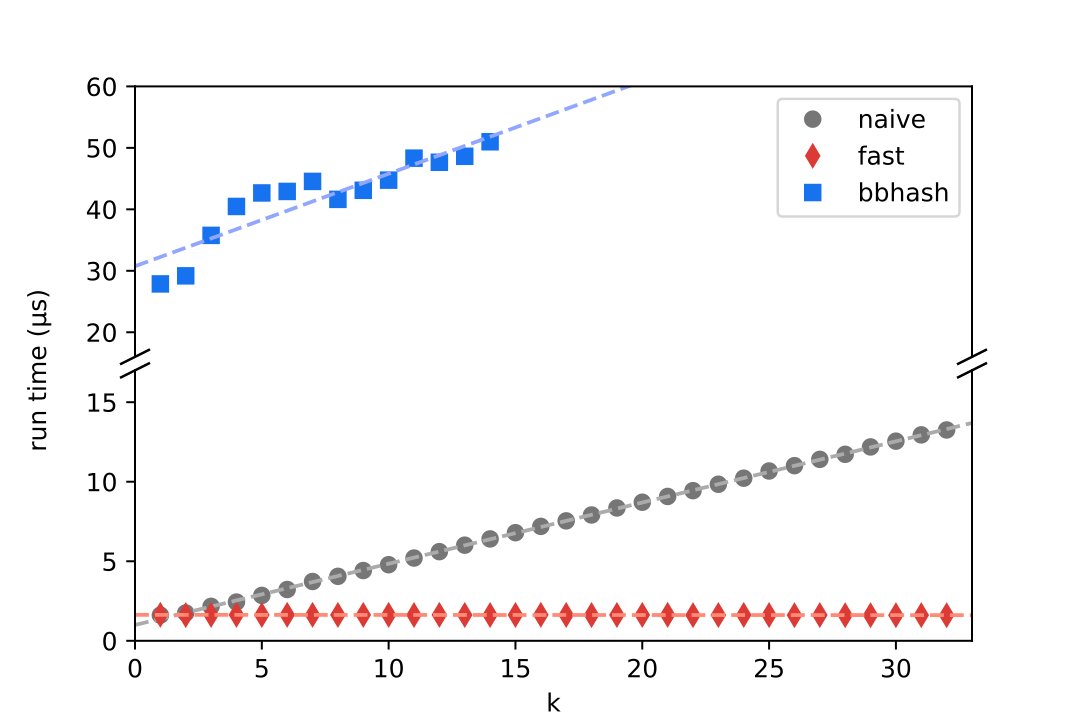
This new preprint describes the method Biotite uses, to decompose sequences into k-mers: The algorithm is simple, fast and its speed is independent of the k-mer length. #Bioinformatics
0
0
5
RT @NaefLuca: 🧬 If you love open source, protein structures and are in NYC, don't miss the one and only @bradyajohnston next week! We'll al…
0
5
0
Biotite now uses to Hatch as build backend. The migration was ridiculously smooth. Thank you, @ThePyPA and plugin developers, for this great build system.
0
0
2
Biotite 0.41.0 has been released and it brings quite a bunch of improvements: full-fletched SDF file format, reading/writing bonds in @buildmodels NextGen PDBx files and more robust structure superimposition. Read the full changelog at #Bioinformatics
0
3
11
With version 0.40.0 Biotite now also provides BinaryCIF support as well as a completely refactored PDBx interface. See the full changelog at #Bioinformatics
Switch from MMTF to BCIF As of July 2, 2024, RCSB PDB will no longer serve PDB data in the MMTF compression format. Users should switch to BinaryCIF (BCIF). Details:
0
0
1
Diving into the Chemical Component Dictionary can be quite entertaining. Thank you, @buildmodels. Me: Can we have 3UQ? Biotite: We have 3UQ in the PDB. 3UQ in the PDB (:
0
0
0
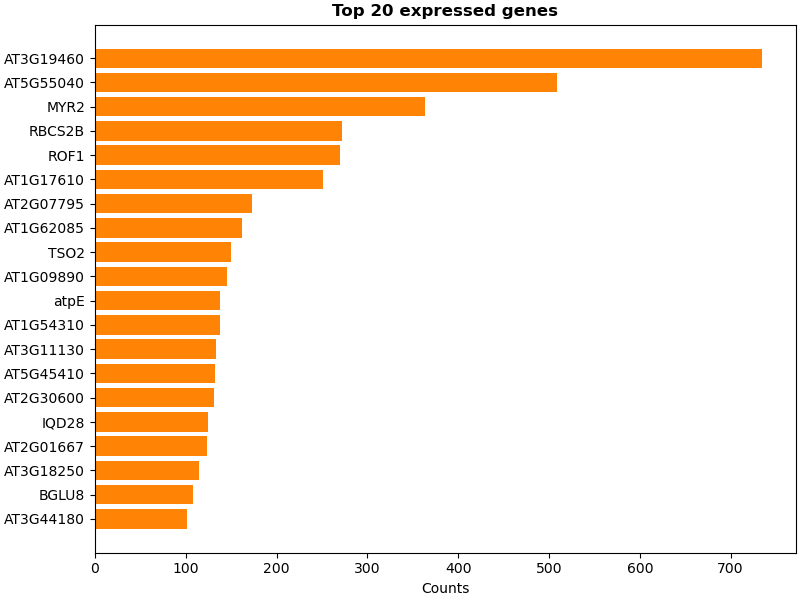
Biotite 0.39 is now released. Beside a multitude of improvements it expands its k-mer matching methods, by supporting minimizers, syncmers or any custom subsetting method. Below is an example using these for gene counting. #Bioinformatics
0
1
4
We have published an update article in BMC Bioinformatics describing major novel features that were added to Biotite in recent years : A big thanks to all co-authors and to all people who contributed to the project in the last 5 years! #Bioinformatics
0
1
1
Here is a small appetizer: The new PubChem API is used to identify all alkane isomers for a given number of carbon atoms. The full example is availabe at #Bioinformatics #Cheminformatics
0
0
0
Working on a bridge between Biotite and @openmm_toolkit to combine powerful MD simulation with convenient data preparation and analysis in a single script. The current state of the development can viewed at #Bioinformatics
0
9
38