Valeria Calvaresi
@Val_Calvaresi
Followers
49
Following
99
Media
0
Statuses
27
Postdoc at @UniofOxford. Interested in the dynamics of viral structures. Like to play with proteins, mass spectrometry & deuterium oxide, better when together.
Oxford
Joined February 2023
We used #nativeMS electrospray ion beam deposition (ESIBD) to make samples for #cryoEM: mass-filtered, soft-landing deposition, cold substrate, in-situ ice growth, clean+cold transfer...we find... @ESIBDLab with many colleagues @OxfordChemistry @KavliOxford
#structuralbiology
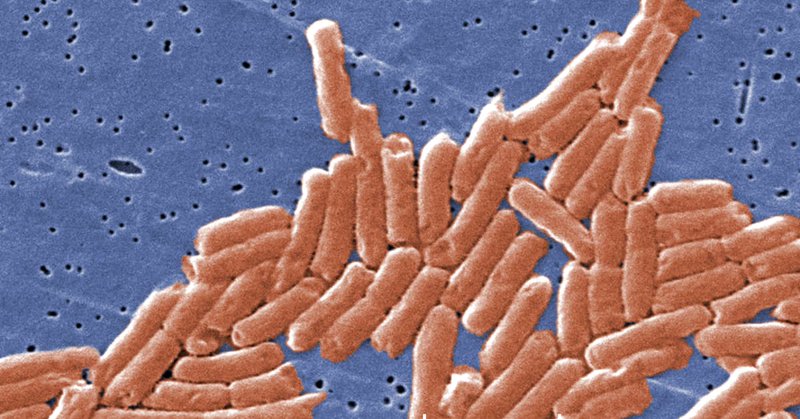
Cryo-EM of soft-landed β-galactosidase: Gas-phase and native structures are remarkably similar https://t.co/HfPSaYRSkd
#bioRxiv
3
31
115
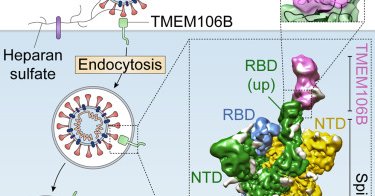
"TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry" @BaggenJim, @DaelemansLab, @TheCrick Peter Cherepanov Read more in @CellCellPress
cell.com
The lysosomal transmembrane protein TMEM106B can serve as an alternative receptor for SARS-CoV-2 entry into ACE2-negative cells. Spike substitution E484D improves spike binding to TMEM106B, enhancing...
0
7
18
Unique opportunity to participate in new structural MS method development for protein-DNA complexes incl. characterization of the nucleic acid 3D structure. We also recruit Postdocs in native MS, ion mobility, HDX-MS and covalent footprinting - happy to chat, DM me. Please RT!
0
2
2
Congrats @wrobel_antoni for a much needed review, and for framing important outstanding questions that suggest directions for future researches👏👏👏
Very excited to share my review on ACE2 binding by SARS-CoV-2 spike, out now @COSB_CRSB! I compare spikes from different sarbecoviruses and review mechanisms that drove spike emergence and evolution during the #COVID19 pandemic from the ACE2 perspective
1
0
0
How can you convince a coronavirus to share its cell entry secrets? Give it something sweet! 🍬 Using #cryoEM & MD, we show how a sugar-based receptor binds to the HKU1 spike and triggers opening via an allosteric mechanism (🧵). https://t.co/Jsm61GjTk8
#glycotime #virology
20
158
447
2x funded PhD in my group at Leeds: pushing frontiers of Structural Mass Spectrometry and application in biology &pharma. (1, EPSRC) ion mobility &AI, (2, MSCA) MS in DNA repair. Intl students can apply - see diff rules for each post. @AstburyCentre @bmss_official @LBMSDG06
1
5
13
Not to say that Lang inferred the basic dynamic mechanisms that underlie protein #HDX and wrote the equations that govern it, which we still use today! This was 10 years before the first protein structures were solved and 20 years before molecular dynamics came on the scene!
1
0
3
By using HDX-MS the group of John Burke @UVicResearch & their collaborators show how different classes of inhibitors alter the conformation of the AGC family protein kinase Akt1, which of interest for future drug design https://t.co/ave9vCkjia
0
9
24
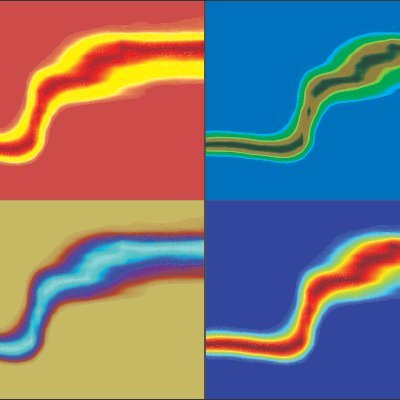
Scientists use #HDX mass spectrometry to describe the evolution of the dynamic structural features of #SARSCoV2 #Spike glycoprotein, and provide insights into the mechanisms of emergence of new virus variants. @Val_Calvaresi @wrobel_antoni @A_Politis
https://t.co/1Jt34j9m2w
0
4
18
Very pleased to see https://t.co/HOISzF6lPz finally published at @NatureComms ! A great effort lead by @Val_Calvaresi at @KingsCollegeLon revealing the dynamics of spikes of variants of concerns with HDX-MS and offering new insights into SARS-CoV-2 evolution.
1
2
12
Here, by #HDXMS we describe the dynamics, energetics and receptor binding of #SARSCoV2 #spike from the Wuhan virus to the omicron variant. Helpful for future HDXMS/structural studies. Thanks to all coauthors, special thanks to @TheCrick - @wrobel_antoni
https://t.co/81ohPx9U4V
0
2
11
Congratulations both, and very well deserved!
We are delighted to give a very special shoutout to Prof. Carol Robinson for receiving the John B. Fenn Award, and to our own, Prof. Brandon Ruotolo for receiving the Beimann Medal!!! We are all so proud of you, Brandon! #ASMS2023 #MassSpec #CIU #UMICH🥳🥳🥳
0
1
13
🧬 70 years ago today, Cambridge theorists Francis Crick and James Watson determined DNA's structure. It changed everything, but the breakthrough was only possible thanks to many talented scientists, including Rosalind Franklin and Maurice Wilkins. Discover more 👇 #OnThisDay
20
186
793
Chromatographic Phospholipid Trapping for Automated H/D Exchange Mass Spectrometry of Membrane Protein–Lipid Assemblies #HDXMS
pubs.acs.org
Lipid interactions modulate the function, folding, structure, and organization of membrane proteins. Hydrogen/deuterium exchange mass spectrometry (HDX-MS) has emerged as a useful tool to understand...
0
0
0