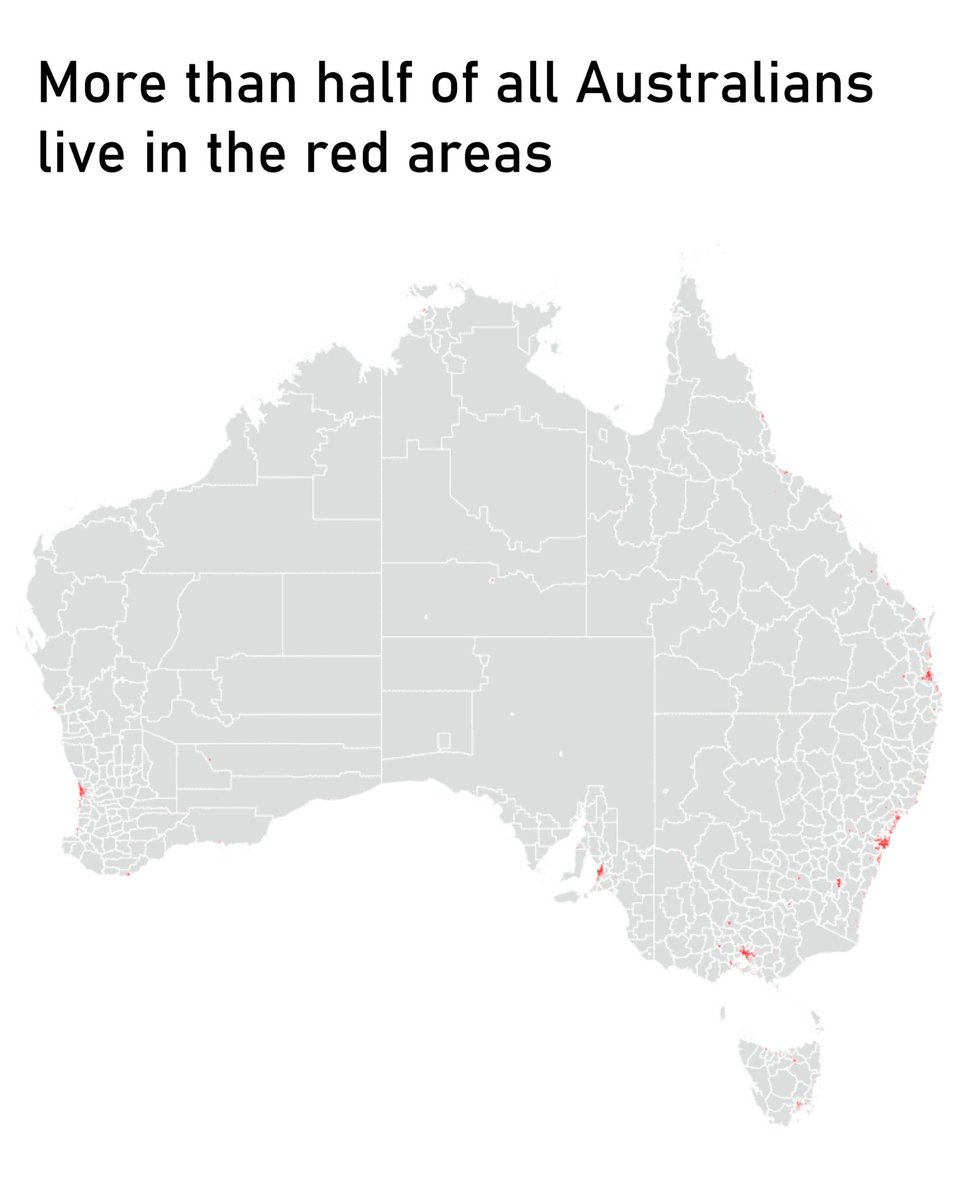
Clive G. Brown
@The__Taybor
Followers
7K
Following
3K
Statuses
22K
RT @amuse: GERMANY: EU regulators think it is too dangerous to simply break a bottle across the bow of a new ship, instead they insist on t…
0
12K
0
RT @EricTopol: Most genome editing work to date has been DNA based. A new paper out today assessed a single dose of epigenetic editing (thr…
0
53
0
RT @EricTopol: Getting your real-time heart rhythm monitored for 10 days now requires an ECG technician to review and notify your doctor of…
0
146
0
@matholomoo No had sensible boots on and a waterproof. Just the shorts. It must have been -12 in the windchill. No he wasn’t Scottish.
1
0
0
RT @ScienceMagazine: Using modern biological tools, researchers engineered #cancer cells with and without specific chromosome abnormalities…
0
154
0
RT @NevilleFChambe1: #RIP #BrianMurphy (1932-2025) Yootha Joyce and Brian Murphy interviewed in February 1979 at the end of the "GEORGE AND…
0
219
0
RT @TsuboiKlein: Thank you @BandaiNamcoUS and @ELDENRING for giving me this gift and congratulating me for being #LetMeSoloHer . I can stil…
0
21K
0
RT @slavov_n: Why does cancer incidence increase with age? Is it the accumulation of mutations? Is it declining immune system? Is it othe…
0
210
0