Priyamvada Acharya
@PriyamvadaA_Lab
Followers
378
Following
204
Statuses
293
Professor @DukeSurgery @dukebiochem @DukeMedSchool @DukeU | Director @DHVI_Struc_Bio | Director @DukeHIVStrucBio | Member @PriyamvadaA_Lab @TheDHVI
Duke University
Joined June 2022
Graduate student Ellie Zhang's cryo-EM structure, beautifully rendered by @swansolive made the cover of Science Translational Medicine. @dukebiochem @TheDHVI @DukeMedSchool @DukeU @AzoiteiLab
This week's issue of #ScienceTranslationalMedicine has arrived! Personalized electric stimulation shows promise in two patients with spinal cord injuries, a study shows that Staph bacteria rapidly evolve resistance to new antibiotic candidates, and more.
0
4
8
RT @AzoiteiLab: Our latest paper was just published in @ScienceTM and highlighted on the cover! 🔬🧪🧬 Congrats to first authors Olivia Swans…
0
6
0
Check out our study on B cell receptor (BCR) structure: The movie below shows the range of motion of the antigen binding fragment (Fab) captured in the cryo-EM dataset. @BhishemThakur @TheDHVI @DukeHIVStrucBio @DukeMedSchool @DukeU
0
2
13
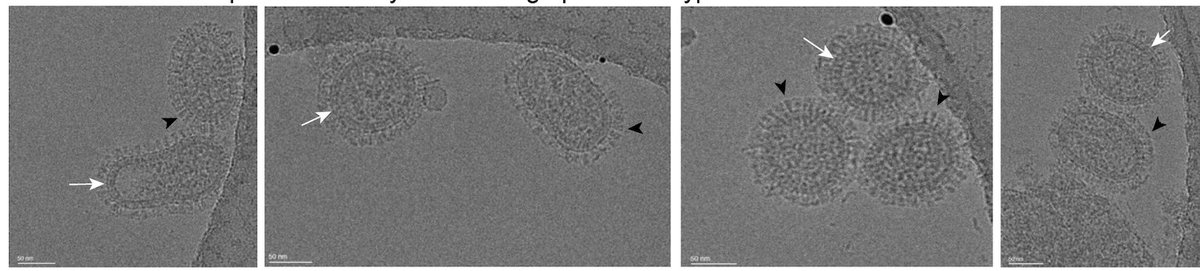
RT @biorxiv_biochem: Structural and antigenic characterization of novel and diverse Henipavirus glycoproteins #bio…
0
2
0
@Aaron_J_May @muralik_lella @HanaXiao93 @Kumar___Ujjwal @kijun_song @katayoun_mn and others not on X. @TheDHVI @dukebiochem @DukeMedSchool @DukeU
0
0
0
RT @RoryCHenderson: Our molecular dynamics simulation based vaccine design paper is out in Nature Communications! We show that studying tr…
0
24
0
Check out special research collection led by @DukeHIVStrucBio affiliated investigators, Maolin Lu, @asharma_lab & @sophie_gobeil Aligned with @DukeHIVStrucBio objectives the collection invites submissions of Original Research, Reviews & Mini-Reviews
0
3
6
It was an honor (and great fun!) to march in the Durham pride parade alongside Duke/DHVI colleagues and the LGBTQ+ community of the larger Durham area. #PrideDurhamNC
@DukeU @TheDHVI @DukeHIVStrucBio
0
1
3
Check out this paper on broadly reactive IgG3 antibodies led by Matt Vukovich @IG_lab. Backstory here is that our collaboration was accelerated by Matt's visit to our lab @DukeU under the Duke Center for HIV Structural Biology Trainee Exchange program @DukeHIVStrucBio
IgG3 #antibodies with exceptional breadth of virus cross-reactivity, with no signs of autoreactivity. Work led by Dr. Matt Vukovich, in collaboration with the groups of @PriyamvadaA_Lab, Giuseppe Sautto, @DannySheward, and others.
0
1
11