MINT database
@MINT_database
Followers
300
Following
358
Statuses
255
MINT is a public database developed at the University of Tor Vergata in Rome and it focuses on experimentally verified protein-protein interactions.
Rome, Lazio
Joined July 2017
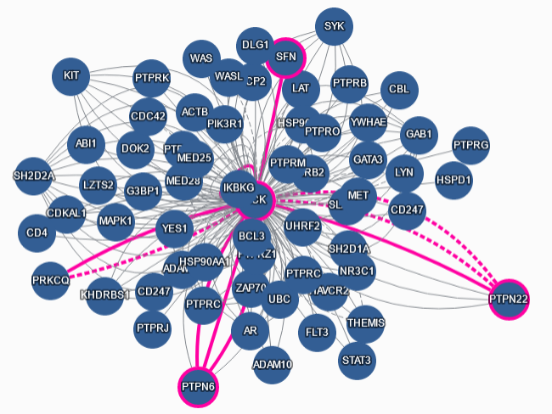
RT @intact_project: Dataset includes 80K point mutations and their effects on interactions. Example shown in pink on this network.
0
1
0
It's time to celebrate!!! @imexconsortium
>1.5 million interactions curated to the@imexconsortiumstandards available for search & download from the IMEx website and Consortium member websites@intact_project,@MINT_database,@uniprot..... Learn more at (Graphic from
0
0
2
RT @pedrobeltrao: new lab preprint - using proteomics and protein co-variation to infer protein association networks across 11 human tissue…
0
43
0
RT @raimondifranc: Our new paper "The landscape of cancer-rewired GPCR signaling axes" is out on Cell Genomics Huge…
0
15
0
We are so pleased that @imexconsortium has been included among the @GlobalBiodata resources! @MINT_database is a founding and an active member of IMEx. IMEx resources have been work jointly to share curation effort and provide a unique high quality protein interactions dataset
We are proud to announce we have been awarded Global Core data resource status by the @GlobalBiodata Coalition. Congratulations to all our member databases: @intact_project, DIP, @MINT_database, MatrixDB, IID, @innatedb, @uniprot, and also @ISBSIB.
0
1
2
RT @guyrcochrane: The GCBRs provide an essential data foundation for the life sciences, both directly to users and as data providers to mor…
0
1
0
RT @imexconsortium: Thank you. So good to see our work on collaboratively producing detailed protein interaction data globally recognised!…
0
2
0
RT @guyrcochrane: Without biodata resources, there is no advance in the life and biomedical sciences. In its 2033 selection, @globalbiodata…
0
4
0
RT @imexconsortium: We are proud to announce we have been awarded Global Core data resource status by the @GlobalBiodata Coalition. Congra…
0
6
0
RT @ELIXIREurope: 🎉 Happy Birthday to ELIXIR! Today we’re celebrating ELIXIR’s 10th anniversary at Brussels 🇧🇪! In the past 10 years, ELIX…
0
28
0
RT @EMBOevents: Sign up now! Abstract submission open until 4 Sep & early bird registration until 20 Sep for EMBO Workshop on Computational…
0
5
0
New interactions curated by MINT team are available via @MINT_database , @intact_project and @imex website after the new IntAct release!!!
📣 Finally, after a year of technical issues, we have managed to release version 244 of IntAct! We are really excited about it, many interesting publications have further extended our coverage. Have a look at
0
4
1
RT @fopsom: Emidio Capriotti highlighting the connection between Rare Diseases and Machine Learning @ELIXIREurope #ELIXIR23. Really interes…
0
3
0
RT @unitorvergata: 📅 3 - 4 maggio | 9.30 | @BiologiaUniRM2 Lo @ucl Cities Partnerships Programme e #unitorvergata presentano il workshop…
0
2
0
RT @unitorvergata: 📆 Martedì, 2 maggio | 9:00 | @BiologiaUniRM2 Lo @UCL - Rome meeting "Bridging the gap between experimentalists, biocur…
0
2
0
RT @biocuration2023: Given the amount of effort that goes into FAIRification of data, it could be considered as a form of curation #biocura…
0
2
0
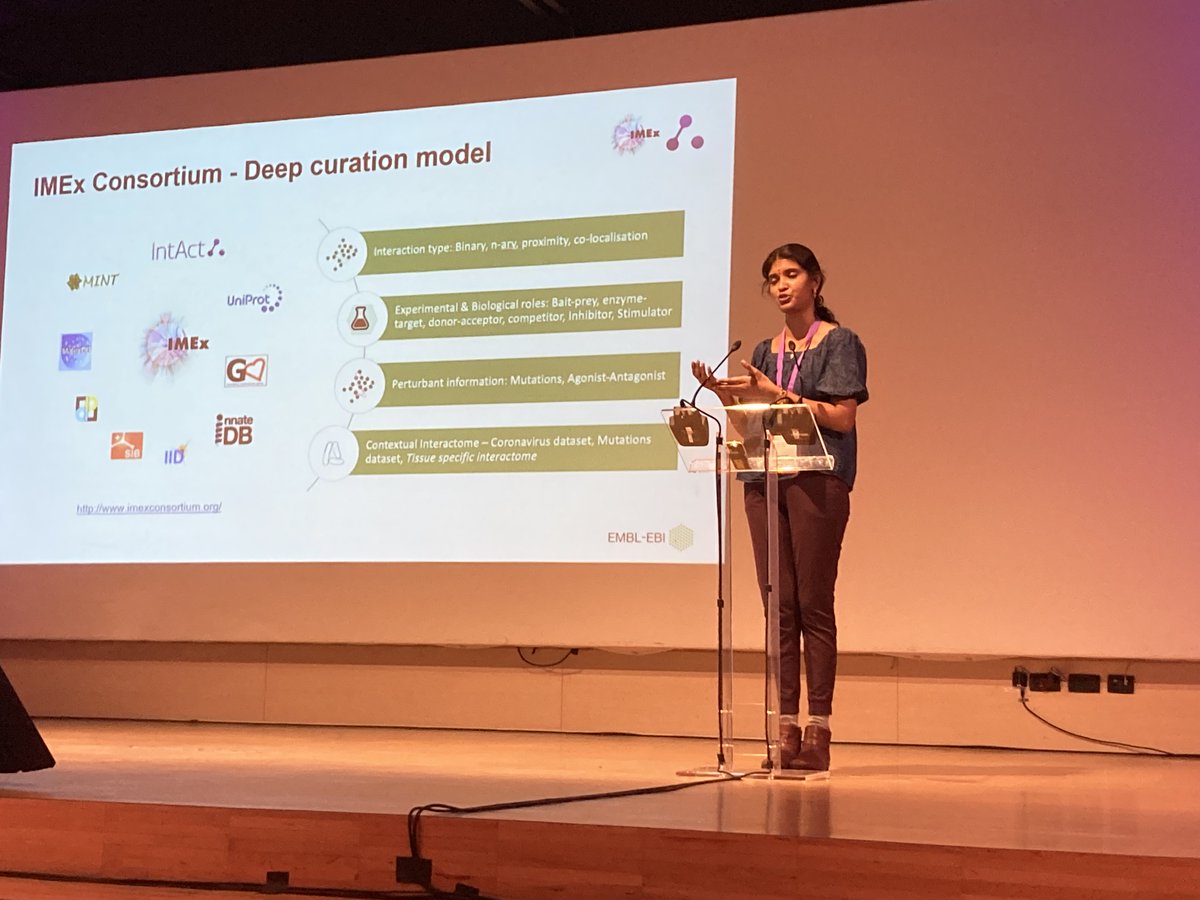
Great talk of Kalpana Panneerselvam from @intact_project team on @imexconsortium’s contextual metadata and interactomes at the Biocuration 2023 day 2. #biocuration2023
0
6
8
RT @biocuration2023: Kicking off the main track of the meeting, Ruth Lovering (@RuthCLovering) is giving a greeting from @biocurator societ…
0
2
0
RT @news4go: Can't recommend this free 2-day #GeneOntology workshop enough!! If you can get to Rome the week after #biocuration2023, sign u…
0
8
0