Linnéa Smeds
@LinneaSmeds
Followers
345
Following
193
Media
26
Statuses
110
Bioinformatician and PhD in evolutionary genetics/conservation genomics, currently doing a PostDoc at Penn State University. Hard core birder when time allows!
State College
Joined May 2020
RT @laurenhennelly1: Sharing this course I'm helping teach at the Smithsonian National Zoo and Conservation Biology institute! 🧬🐺🐻🧬🐦. It's….
smconservation.gmu.edu
This 10-day residential course for graduate students and professionals provides a survey of the concepts, methods, and software used in conservation genomics research.
0
33
0
RT @MakovaLab: The @MakovaLab had a great time #SMBE2024 presenting & learning about cutting-edge research and enjoying #Mexican hospitalit….
0
3
0
RT @judithmank: Cover image initiative at Evolution Letters! We are looking for artists who would like to work with authors of Evolution Le….
0
34
0
RT @JesperBoman: MEIOTIC DRIVE AND THE EVOLUTION OF CHROMOSOMES. In 2001, Pardo-Manuel de Villena and Sapienza observed that positions of c….
0
23
0
RT @lisaklas: Are you interested in how co-evolution shapes the genomes of bacterial symbionts and their hosts? Open PhD position in my lab….
0
21
0
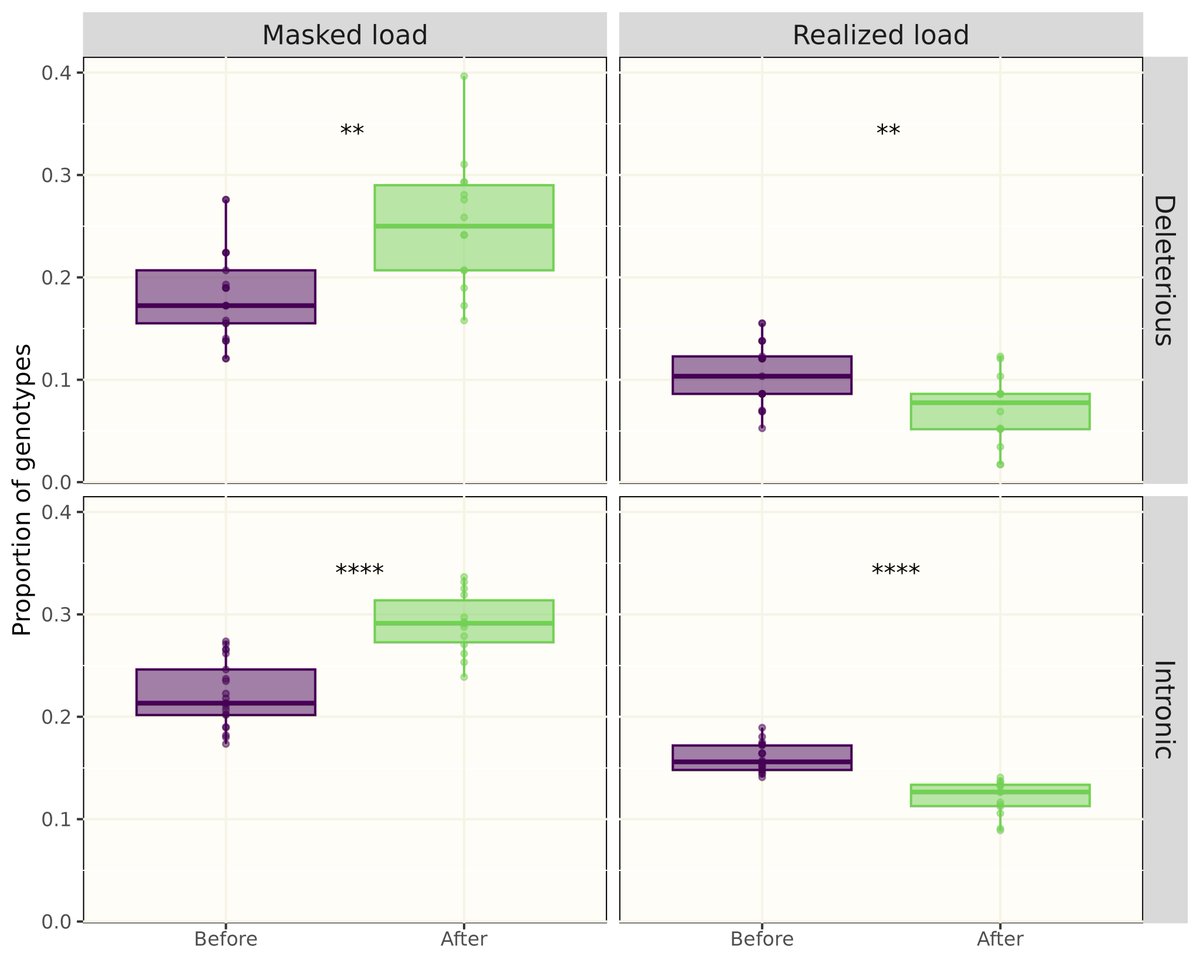
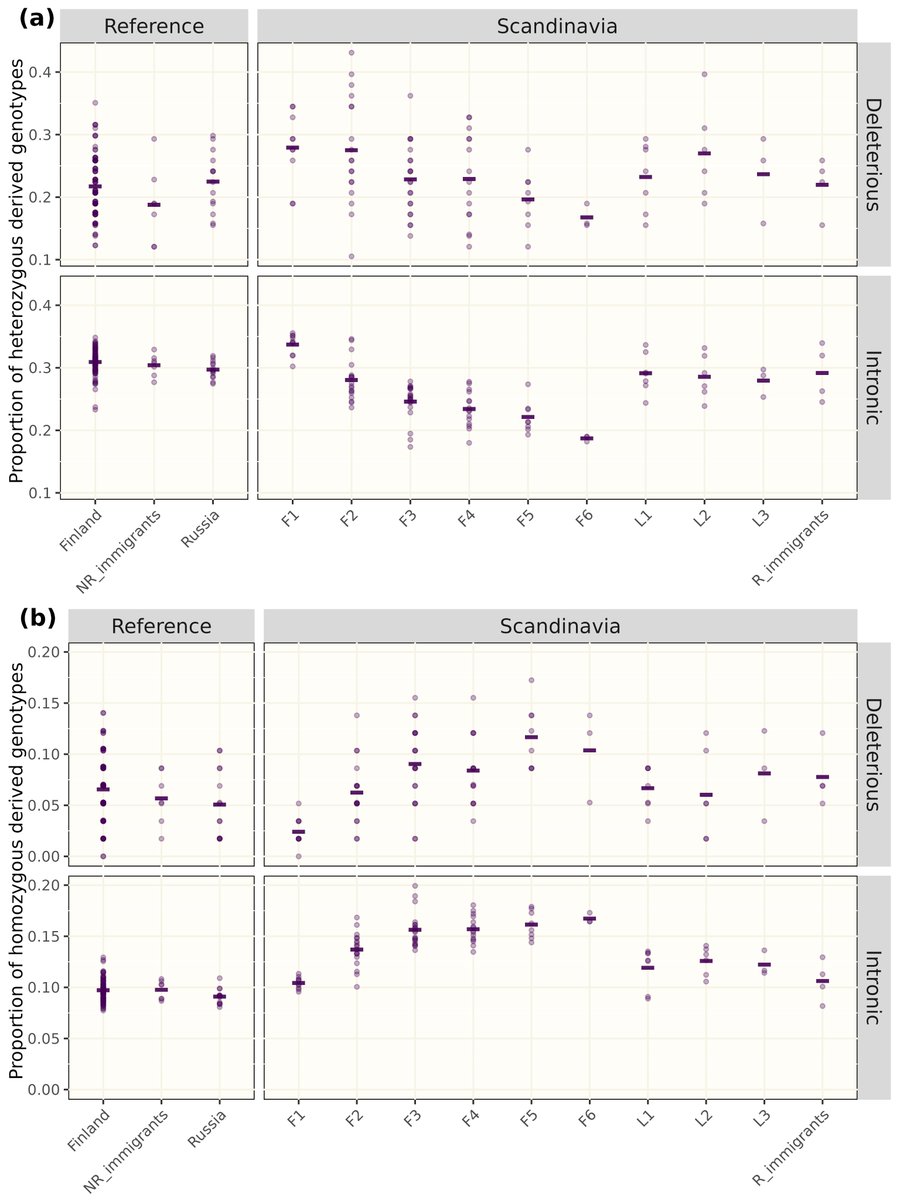
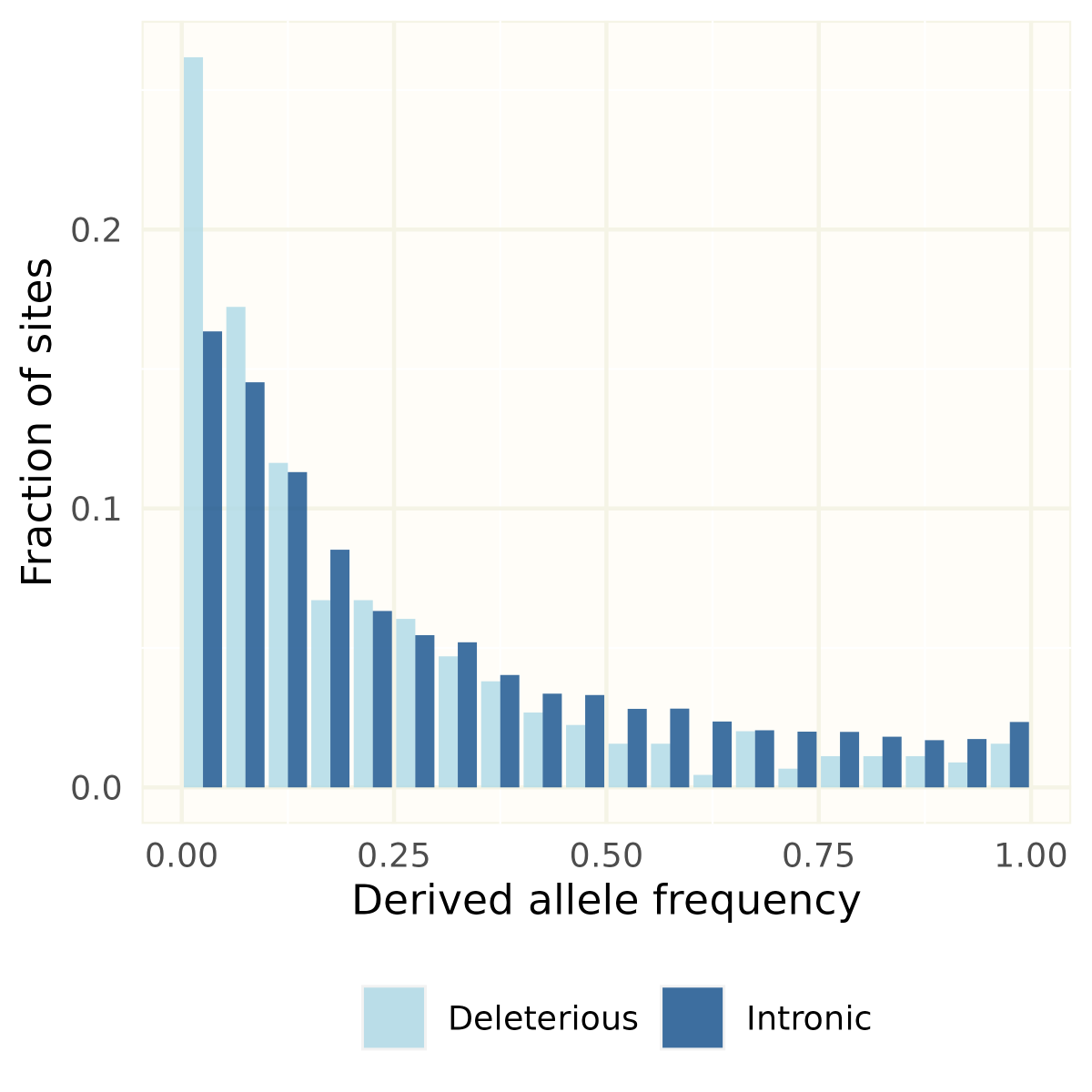
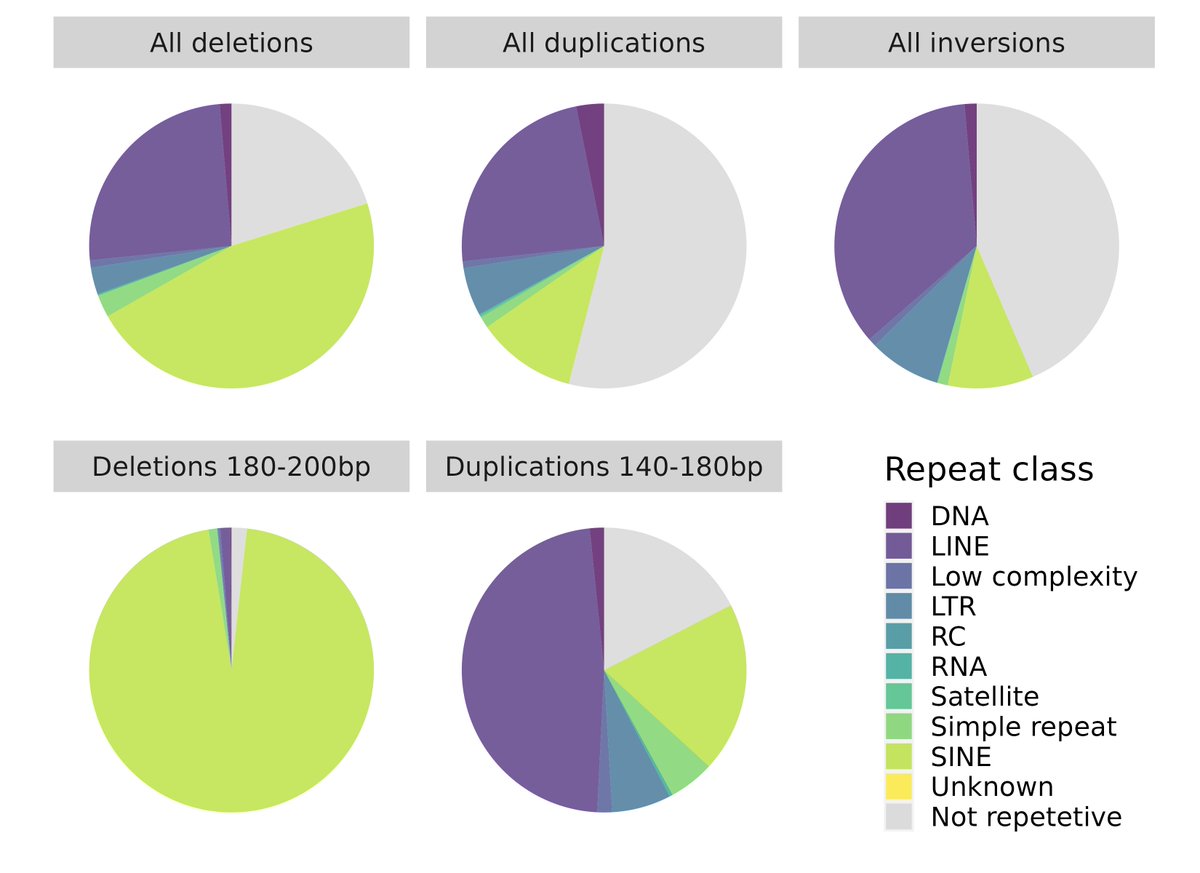
RT @EvolAppJournal: Structural variation and genetic load in inbred Scandinavian wolves, and how they are affected by a genetic rescue ev….
0
5
0
Come work with great people at @EvoBioUppsala (my favorite workplace since 2009😊).
I am officially looking for a PhD student to work with me on questions regarding the evolution of lichens algal partners. I would be really grateful if you can help me spread the add as far as possible. #PhD #lichens #photobionts #jobsearch
0
3
6
Last Friday I defended my thesis about #Conservation #genomics in Scandinavian #Wolves at @UU_University. I enjoyed every second of the day, especially the discussion with my opponent Giorgio Bertorelle. A great end to an era in my life. Thanks to everyone who's been part of it♥️
12
2
70
Our paper about structural variants in inbred Scandinavian #wolves just came out in @EvolAppJournal! See 🧵 below on how we used them to investigate genetic load⬇ (1/8).
4
27
67