Nikel SEM Group
@LabNikel
Followers
2K
Following
1K
Statuses
799
Building synthetic metabolisms in bacteria for a sustainable future @DTUBiosustain · Views are our own + #Pseudomonas’ 🧬 ·
Copenhagen, Denmark
Joined August 2018
RT @PabloINik: CIFR (Clone–Integrate–Flip-out–Repeat): A toolset for iterative #genome and pathway #engineering of Gram-negative #bacteria—…
0
10
0
RT @MichelePartipi3: Amazing tool for multiple cycles of genetic engineering of Gram negative bacteria developed by @FilFederici and France…
0
1
0
RT @PabloINik: Leveraging engineered #Pseudomonas putida minicells for bioconversion of organic acids into short-chain methyl ketones by @K…
0
11
0
Touching on new bugs, here @enricoorsi and @dellavallesimo led the work for creating a new CRISPR/Cas toolkit for the emerging C1 platform C. necator, showing unprecedented efficiencies and turnover times for gene deletions
1
0
3
And in this chapter, w/ @DeMaria_SynBio they outline use of L-threonine aldolases (LTAs) from E.coli and P.putida to efficiently synthesize 4-F-L-threonine in vitro + protocols for adapting these activities to produce other fluorometabolites in vivo 10.1016/bs.mie.2024.02.016
1
0
1
Going back to P. putida, this minireview w/ @vdlorenzo_CNB and @PabloINik traces the evolution of KT2440 from a model organism for biodegradation studies to a SynBio #chassis, highlighting its adaptability and importance in various applications
1
0
1
When talking #biosensors, it is important to understand that their construction and validation is not easy. Here, @enricoorsi & @mirbachh demonstrated an improved end-to-end workflow for building a suite of sensors to measure glyoxylate and glycolate
1
0
2
Whereas here @Javi_HernanSan developed a whole-cell #biosensor for co-culture setups that integrates #fluorescent reporters and synthetic auxotrophies, enabling detection of multiple molecules, including plastic degradation and environmental analysis.
1
0
1
Talking about #biosensors, here @FiorellaMasotti developed one for #glyphosate detection by engineering A. tumefaciens with an optimized phnG promoter, enabling detection in the 0.25–50 μM range in soil and water samples
1
0
1
@FavoinoGiusi demonstrates here how to increase malonyl-CoA availability in P. putida using biosensor-informed engineering, and enhancing polymer production through a non-canonical pathway to boost the output of malonyl-CoA-dependent biosynthetic processes
1
0
1
Here are the fluoro masters @DeMaria_SynBio & @ManuelNietoDom1 presenting a protocol for synthesizing 4-F-L-threonine using L-threonine aldolases from E. coli or P. putida, highlighting the potential of F-amino acids for new chemistries and applications
1
0
1
Then @enricoorsi & @Javi_HernanSan reason how by using noncanonical redox cofactors in C1-bioprocesses can enhance product yields and enable challenging redox reactions, overcoming limitations of natural cofactors, contributing to a circular carbon economy
1
0
2
We start with a review led by @erayubozkurt describing strategies for isolating, creating, & characterizing enzyme variants optimized for bioproduction, including high-throughput screening and automated systems for developing novel biocatalysts
1
0
1
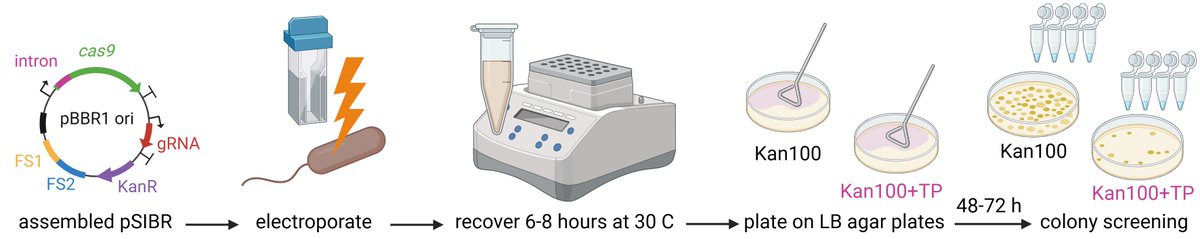
New paper from the group developing a cutting edge genome editing tool for Cupriavidus necator! With @enricoorsi, @copa_bio, @dellavallesimo leading the work and including @NicoC_MicSynBio @PabloINik Chek below form more info!
We developed a brand new #CRISPR toolkit for #Cupriavidus #necator, a biotechnological platform for CO2 (and derivatives) valorization. The system is based on self-splicing introns and allows us to get mutants within 48h and with efficiencies up to 80% Check the thread below👇
0
6
33