Kimberley Billingsley
@KimberleyBill10
Followers
593
Following
4K
Media
32
Statuses
545
Head of the Applied Neurogenomics group at @NIH @NIA @CARD | 🧠🧬 Using Long Reads to better understand the genomics of ADRD | Views/tweets my own
Joined October 2018
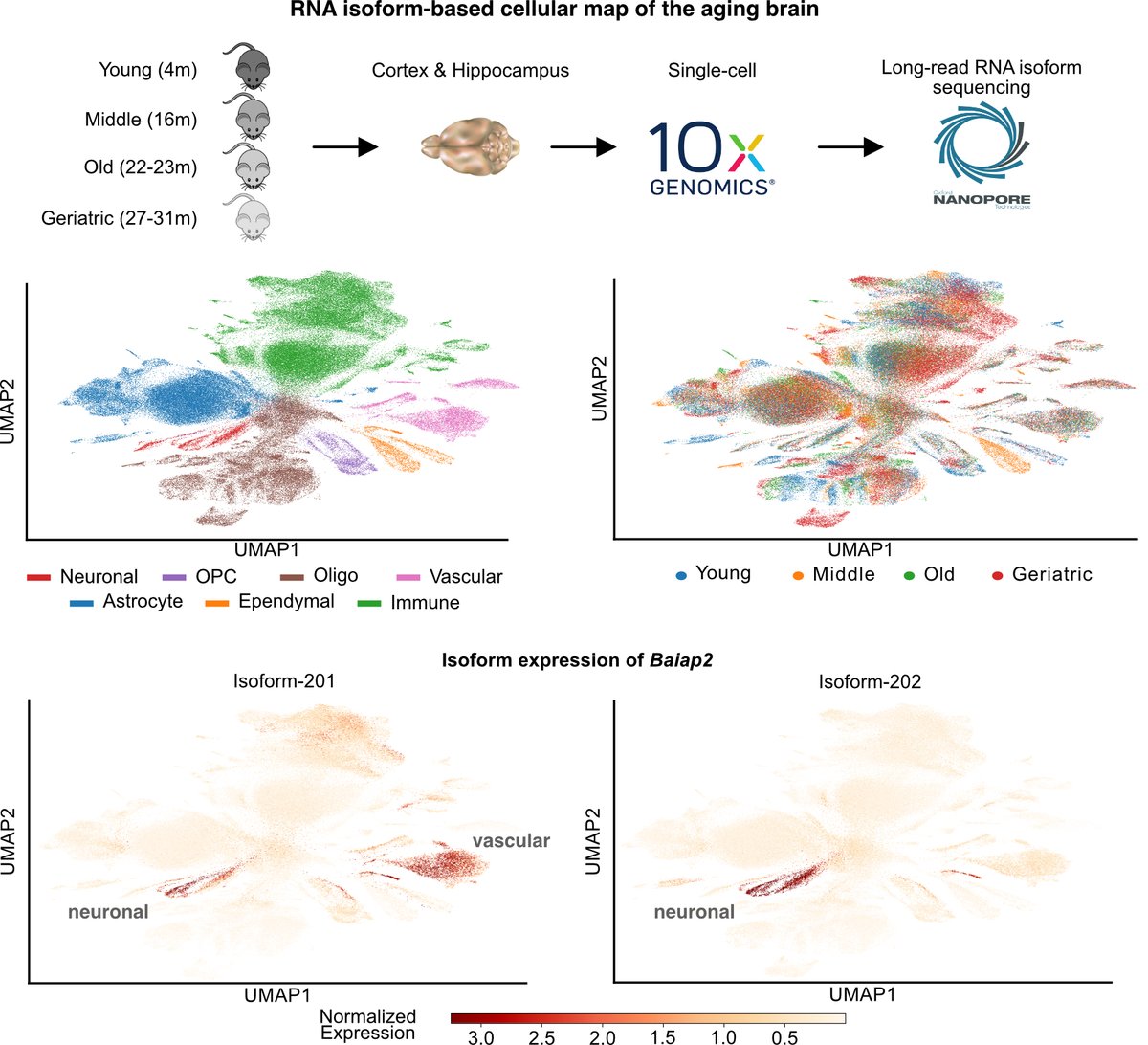
Excited to share our work with Mark Cookson and @senergystic labs. We combine #SingleCell and #Nanopore sequencing to profile #RNA isoforms from young to geriatric mouse brains. We uncover #aging changes in transcription, processing, and splicing that also impact coding potential
Cell-specific RNA isoform remodeling in the aging mouse brain https://t.co/oJr5MnBVVv
#biorxiv_genomic
2
13
36
Chile spotted! SCA 4 not only in Sweden, also in the south of the world in admixed population, with @ONT and @KimberleyBill10 #CARD team. Check it out “Identification of GGC Repeat Expansions in ZFHX3 among Chilean Movement Disorder Patients”
movementdisorders.onlinelibrary.wiley.com
Background Hereditary ataxias are genetically diverse, yet up to 75% remain undiagnosed due to technological and financial barriers. The GGC repeat expansion in ZFHX3, responsible for spinocerebell...
1
4
11
In @NatureGenet we report that base editing of trinucleotide repeats (TNRs) reduces somatic repeat expansions in Huntington’s disease (HD) and Friedreich’s ataxia (FRDA)—in patient-derived cells and in vivo—a collaboration with the Mouro Pinto lab. https://t.co/J6MhwEAgoB 1/14
17
133
561
0
2
11
Come join us at our 7th #bioinformatics #hackathon @BCM_HGSC @BCMFromtheLabs around Structural Variants, Graph genomes & many related topics: https://t.co/QDdqGvHRYY Groups will work on interesting topics that will be published in F1000. Registration closes 10th of Aug!
1
10
11
Excited & very lucky to be part of the Gates Sr. AD Fellowship ✨ Using CARD’s long-read epigenetic data 🧬 to build better predictive models of ADRD & making all models & methods openly available (open science FTW!🔓). Looking forward to collaborating with this awesome group!
Meet Kimberley Billingsley, PhD! 🎉 As a Gates Sr. AD Fellow, she will use high-resolution methylation data to develop predictive models for ADRD, identify epigenetic signatures for better risk stratification and early detection, and establish accurate biomarkers. #ADRD
1
3
24
Long reads @nanopore @PacBio can provide insights into epigenetics. We (@fuyilei96 @timp0) summarize the latest #Bioinformatics approaches to analyze methylation & why this matters! Out now @NatureRevGenet
https://t.co/rbjJcDQOrP
@BCM_HGSC @bcmgenetics @RiceCompSci @JohnsHopkins
1
74
287
Collaboration made the impossible possible: we identified the first SCA4 individuals outside Northern Europe—right here in #Chile through @nanopore Speaking a common scientific language @KimberleyBill10 breaks barriers for underserved patients! #RareDisease #Genetics
Identification of GGC Repeat Expansions in ZFHX3 Among Chilean Movement Disorder Patients https://t.co/0DrdPYyGxQ
#medRxiv
0
5
14
Delighted to comment on the paper explaining the role of a new GBA variant in Parkinson's risk in Africa in Nature Structural and Molecular Biology by @pilaralvjer
1
3
8
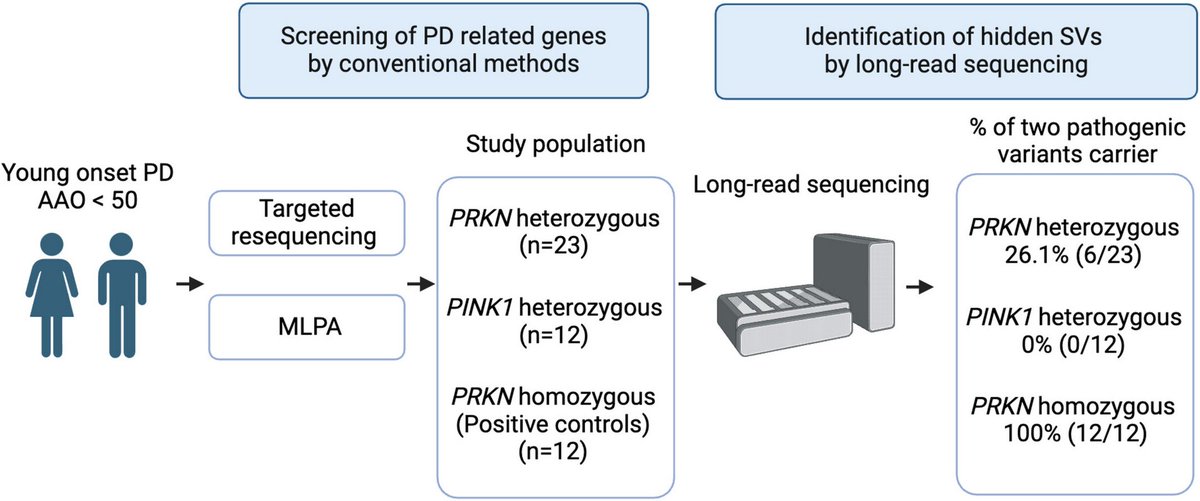
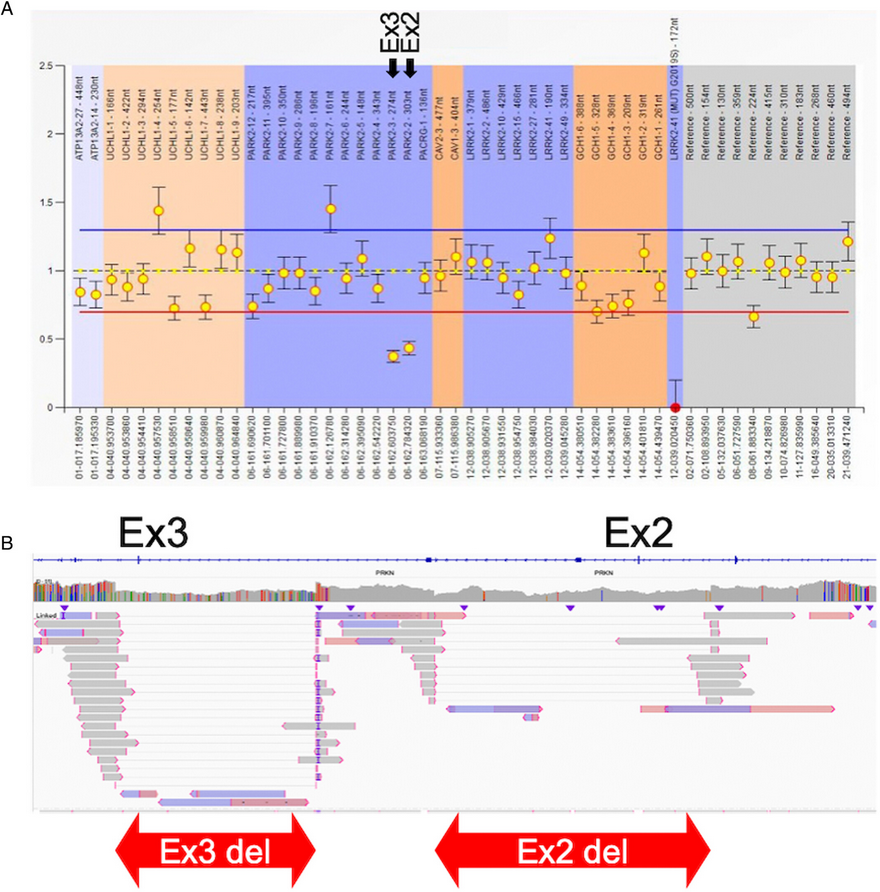
Researchers using @nanopore long-read sequencing effectively identify pathogenic structural variants in the PRKN locus that were missed by conventional methods; They propose that it should be considered for unresolved early onset #Parkinsons cases https://t.co/VWwi7nfBy9
1
6
18
Excitingly, lots more data to be released in 2025, and we’re eager to expand this framework to include disease cohorts next! 🎉✨ @cornelisblauw @mike_nalls @SingletonNeuro #LongReadSequencing #Neurogenomics #DiversityInGenomics #OpenScience
0
1
2
12/ Finally, this study is part of the CARD long-read sequencing initiative, which aims to sequence thousands of Alzheimer’s Disease and Related Dementias samples from diverse populations to uncover the genetic and epigenetic drivers of these diseases on a global scale.🌍
1
1
1
11/ 📢 Check out the preprint here: https://t.co/vYcbGIJWXU We’d love to hear your feedback, ideas, and collaboration opportunities. 🚀
biorxiv.org
Structural variants (SVs) drive gene expression in the human brain and are causative of many neurological conditions. However, most existing genetic studies have been based on short-read sequencing...
1
1
2
10/ 🔬 Open Science! All NAPU outputs (SVs, SNVs, assemblies, mapped BAMs, etc.) will be shared on AnVIL! 🚀The data is currently being uploaded, and we’ll keep you posted when it’s live🔑Note: You’ll need prior dbGaP access to datasets phs001300 and phs000979 to access the data
1
1
2
9/ This resource lays the groundwork for future studies, enabling: - Investigations into neurodegenerative disease mechanisms (e.g., ADRD) - Insights into ancestry-specific genetic variation Understanding how SVs shape gene expression and DNA methylation
1
1
1
8/🧩 Why is this important? Our findings emphasize the important role of SVs in brain biology. By including ancestrally diverse samples, we’ve built a foundation for inclusive research into how gene regulation impacts neurological diseases. 🌍🧠
1
1
2
7/ More highlights: - 34,087,867 small variants 🧬 - Methylation profiles at 28+ million CpG sites 🔬 - This provides a unique opportunity to study the genetic and epigenetic landscape of the brain at an unparalleled resolution.
1
1
1
6/ We also uncovered haplotype-specific DNA methylation at millions of CpG sites, identifying cis-acting SVs with significant regulatory impacts. LRS highlights complex regulatory mechanisms that SRS can't resolve. 🌌
1
0
0
5/ By integrating matched gene expression data, we performed QTL analyses, showing how SVs impact gene expression in the frontal cortex🧠 Over 50% of these top nominated causal SVs were missed by SRS, including a key deletion at the NLRP2 locus linked to neuroinflammation. 🔑🔥
1
0
0
4/ What we found: - ~250k SVs 🧩 - Locally phased assemblies covering 95% of protein-coding genes in GRCh38 🧬 - Many of these SVs are missed by short-read sequencing (SRS)
1
0
0