Katie Yost
@KatieEYost
Followers
619
Following
295
Statuses
50
NCI K00 Postdoctoral Fellow, @JswLab, @WhiteheadInst | PhD @HowardYChang Lab, @stanfordcbio
Cambridge, MA
Joined January 2017
RT @dianyang1117: Excited to announce the Yang lab officially opens at @Columbia University. We are interested in developing innovative sin…
0
82
0
@EmilyAshkin @NIH @theNCI @LabWinslow @CancerHacker @JswLab @WhiteheadInst Congrats Emily!! So excited to have you around!
0
0
0
RT @facts___matter: So excited to share our work out in @NatImmunol !!! We analyzed antigen-specific CD8 T cells differentiating down effec…
0
105
0
@vangalenlab @KyleRomine12 @NatImmunol Hi Peter - our data suggests that transgenic TCR systems (typically high avidity) would be biased towards a single differentiation trajectory but that endogenous polyclonal T cells have heterogeneous differentiation trajectories driven by differences in TCR avidity.
0
0
2
@BingfeiY @KECKSchool_USC @uscnorris Congratulations Bingfei! Looking forward to the inspiring science to come from your lab!
1
0
1
RT @BingfeiY: 🎉My turn! Super excited to join @KECKSchool_USC @uscnorris as an Assistant Professor in early 2023. My lab will decode inter…
0
13
0
Very grateful to work with fantastic co-authors and collaborators including @BenceDaniel2 Sunnie Hsiung @HowardYChang @FW14B_Red5 @Satpathology
0
0
0
Beautiful work on spatial mapping of T cell clones!
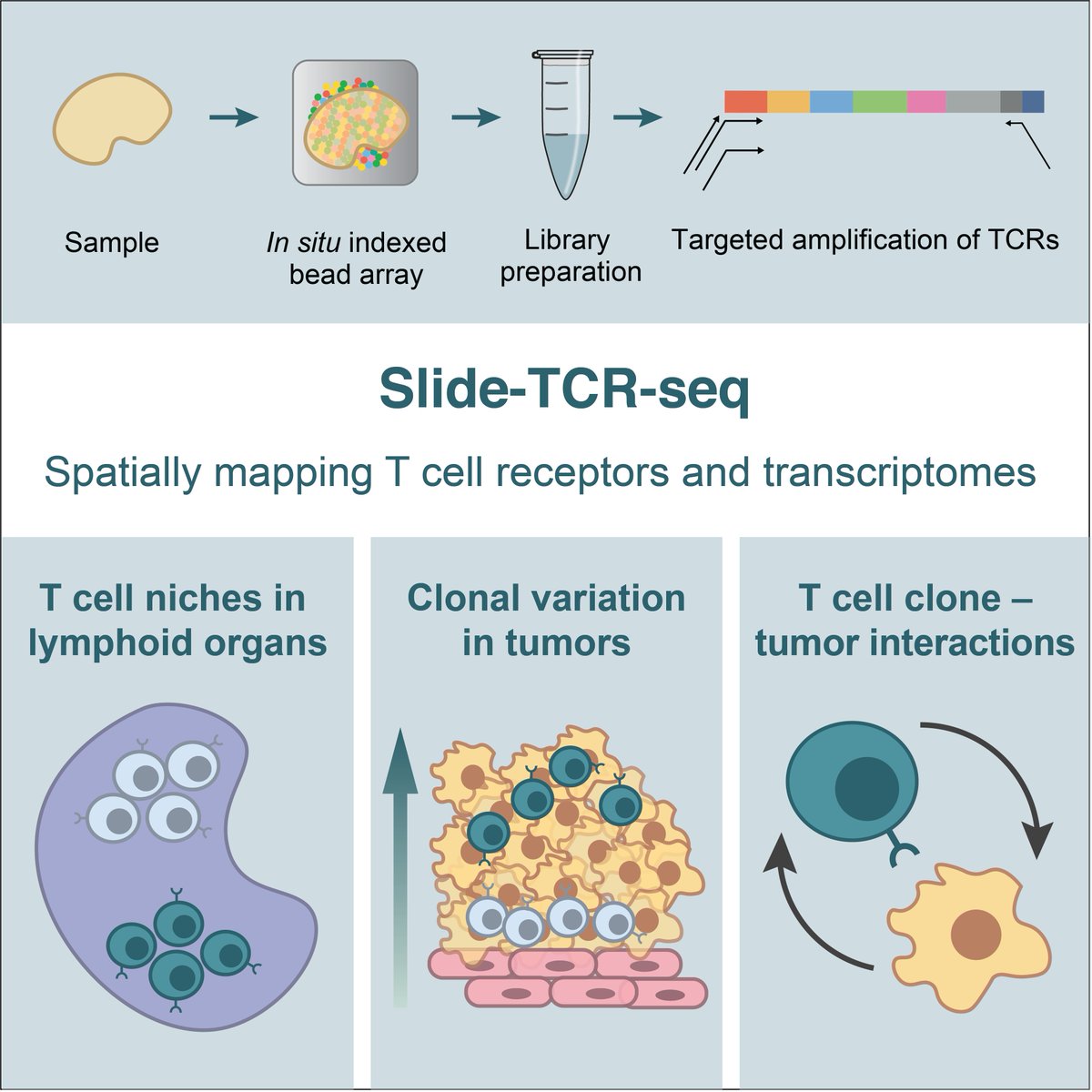
Presenting Slide-TCR-seq: our method for measuring the transcriptomes of cells & T cell receptor (TCR) sequences in situ! This collaboration w/ Bryan Iorgulescu, Shuqiang Li, Catherine Wu, and @insitubiology led to some cool findings... 1/8
0
0
8
RT @josephmreplogle: I'm thrilled to share our latest work performing genome-scale Perturb-seq screens to explore a remarkable breadth of q…
0
156
0
Our preprint on divergent clonal differentiation trajectories during T cell exhaustion is now on bioRxiv! We investigate whether all antigen-specific T cell clones follow the same differentiation trajectory during chronic viral infection and find heterogeneous clonal behaviors.
Excited to share our preprint on clonal T cell differentiation trajectories of T cell exhaustion. Great collaboration with @KatieEYost @HowardYChang @FW14B_Red5 @Satpathology and others @StanfordMed.
1
16
70
RT @NatureRevCancer: NEW #ResearchHighlight! @kinglhung, @KatieEYost, Xie @549nm et al. @HowardYChang show that extrachromosomal DNAs (#ecD…
0
10
0
RT @kinglhung: Today we release in @biorxivpreprint a method for isolating megabase-sized, oncogenic extrachromosomal DNA from human cancer…
0
29
0
@ara_latifkar @kinglhung @Nature @HowardYChang @549nm Thank you @ara_latifkar ! Besides high levels of transcription I think not a lot known… yet :)
0
0
4
And excited to see work from @_eunhee_yi_ @roelverhaak et al using live cell imaging to visualize transcriptionally active ecDNA hubs, also out today! (3/3)
0
0
6
So grateful for the opportunity to present my PhD work at Stanford Cancer Biology retreat!
Congratulations to @KatieEYost on receiving the 2021 Denise A. Chan Outstanding Thesis Award for her work on clonal replacement of tumor T cells upon PD-1 blockade AND ecDNA hub-driven cooperative oncogene expression 🦀🦀🦀
3
1
52